Exploring the Microdiversity Within Marine Bacterial Taxa: Toward an Integrated Biogeography in the Southern Ocean
- PMID: 34335536
- PMCID: PMC8317501
- DOI: 10.3389/fmicb.2021.703792
Exploring the Microdiversity Within Marine Bacterial Taxa: Toward an Integrated Biogeography in the Southern Ocean
Abstract
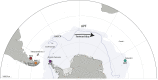
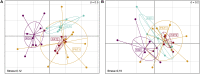
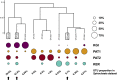
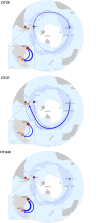
Most of the microbial biogeographic patterns in the oceans have been depicted at the whole community level, leaving out finer taxonomic resolution (i.e., microdiversity) that is crucial to conduct intra-population phylogeographic study, as commonly done for macroorganisms. Here, we present a new approach to unravel the bacterial phylogeographic patterns combining community-wide survey by 16S rRNA gene metabarcoding and intra-species resolution through the oligotyping method, allowing robust estimations of genetic and phylogeographic indices, and migration parameters. As a proof-of-concept, we focused on the bacterial genus Spirochaeta across three distant biogeographic provinces of the Southern Ocean; maritime Antarctica, sub-Antarctic Islands, and Patagonia. Each targeted Spirochaeta operational taxonomic units were characterized by a substantial intrapopulation microdiversity, and significant genetic differentiation and phylogeographic structure among the three provinces. Gene flow estimations among Spirochaeta populations support the role of the Antarctic Polar Front as a biogeographic barrier to bacterial dispersal between Antarctic and sub-Antarctic provinces. Conversely, the Antarctic Circumpolar Current appears as the main driver of gene flow, connecting sub-Antarctic Islands with Patagonia and maritime Antarctica. Additionally, historical processes (drift and dispersal limitation) govern up to 86% of the spatial turnover among Spirochaeta populations. Overall, our approach bridges the gap between microbial and macrobial ecology by revealing strong congruency with macroorganisms distribution patterns at the populational level, shaped by the same oceanographic structures and ecological processes.
Keywords: Antarctic Circumpolar Current; Antarctic Polar Front; Microbial Conveyor Belt; Minimum Entropy Decomposition; Southern Ocean; Spirochaeta; microdiversity; phylogeography.
Copyright © 2021 Schwob, Segovia, González-Wevar, Cabrol, Orlando and Poulin.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Adamack A. T., Gruber B. (2014). PopGenReport: simplifying basic population genetic analyses in R. Methods Ecol. Evol. 5 384–387.
-
- Almasia R., Carú M., Handford M., Orlando J. (2016). Environmental conditions shape soil bacterial community structure in a fragmented landscape. Soil Biol. Biochem. 103 39–45. 10.1016/j.soilbio.2016.08.004 - DOI
-
- Arkhipkin A. I., Middleton D., Sirota A., Grzebielec R. (2004). The effect of Falkland current inflows on offshore ontogenetic migrations of the squid Loligo gahi on the southern shelf of the Falkland Islands. Estuar. Coast. Shelf Sci. 60 11–22. 10.1016/j.ecss.2003.11.016 - DOI
-
- Assis J., Tyberghein L., Bosch S., Verbruggen H., Serrão E. A., De Clerck O. (2018). Bio−ORACLE v2. 0: extending marine data layers for bioclimatic modelling. Glob. Ecol. Biogeogr. 27 277–284. 10.1111/geb.12693 - DOI
-
- Astorga A., Oksanen J., Luoto M., Soininen J., Virtanen R., Muotka T. (2012). Distance decay of similarity in freshwater communities: do macro−and microorganisms follow the same rules? Glob. Ecol. Biogeogr. 21 365–375. 10.1111/j.1466-8238.2011.00681.x - DOI
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

