High-Density Blood Transcriptomics Reveals Precision Immune Signatures of SARS-CoV-2 Infection in Hospitalized Individuals
- PMID: 34335605
- PMCID: PMC8322982
- DOI: 10.3389/fimmu.2021.694243
High-Density Blood Transcriptomics Reveals Precision Immune Signatures of SARS-CoV-2 Infection in Hospitalized Individuals
Abstract
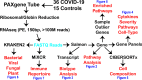
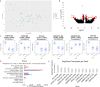
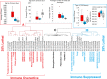
The immune response to COVID-19 infection is variable. How COVID-19 influences clinical outcomes in hospitalized patients needs to be understood through readily obtainable biological materials, such as blood. We hypothesized that a high-density analysis of host (and pathogen) blood RNA in hospitalized patients with SARS-CoV-2 would provide mechanistic insights into the heterogeneity of response amongst COVID-19 patients when combined with advanced multidimensional bioinformatics for RNA. We enrolled 36 hospitalized COVID-19 patients (11 died) and 15 controls, collecting 74 blood PAXgene RNA tubes at multiple timepoints, one early and in 23 patients after treatment with various therapies. Total RNAseq was performed at high-density, with >160 million paired-end, 150 base pair reads per sample, representing the most sequenced bases per sample for any publicly deposited blood PAXgene tube study. There are 770 genes significantly altered in the blood of COVID-19 patients associated with antiviral defense, mitotic cell cycle, type I interferon signaling, and severe viral infections. Immune genes activated include those associated with neutrophil mechanisms, secretory granules, and neutrophil extracellular traps (NETs), along with decreased gene expression in lymphocytes and clonal expansion of the acquired immune response. Therapies such as convalescent serum and dexamethasone reduced many of the blood expression signatures of COVID-19. Severely ill or deceased patients are marked by various secondary infections, unique gene patterns, dysregulated innate response, and peripheral organ damage not otherwise found in the cohort. High-density transcriptomic data offers shared gene expression signatures, providing unique insights into the immune system and individualized signatures of patients that could be used to understand the patient's clinical condition. Whole blood transcriptomics provides patient-level insights for immune activation, immune repertoire, and secondary infections that can further guide precision treatment.
Keywords: COVID-19; RNAseq; SARS-CoV-2; blood transcriptomics; immune cell deconvolution; immune repertoire; interferon response; secondary infections.
Copyright © 2021 Prokop, Hartog, Chesla, Faber, Love, Karam, Abualkheir, Feldmann, Teng, McBride, Leimanis, English, Holsworth, Frisch, Bauss, Kalpage, Derbedrossian, Pinti, Hale, Mills, Eby, VanSickle, Pageau, Shankar, Chen, Carcillo, Sanfilippo, Olivero, Bupp and Rajasekaran.
Conflict of interest statement
JP performed a summer sabbatical in 2020 for AbbVie Inc, receiving hourly pay. RK, NA, BF, and LT were employees of Ambry Genetics during this study. NK is an advisory board member for Horizon pharmaceuticals and Pharming Healthcare. None of these listed conflicts were directly related to the current work. The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

