Development of a Four-mRNA Expression-Based Prognostic Signature for Cutaneous Melanoma
- PMID: 34335689
- PMCID: PMC8320537
- DOI: 10.3389/fgene.2021.680617
Development of a Four-mRNA Expression-Based Prognostic Signature for Cutaneous Melanoma
Abstract
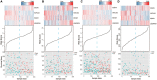
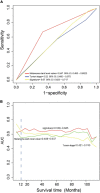
We aim to find a biomarker that can effectively predict the prognosis of patients with cutaneous melanoma (CM). The RNA sequencing data of CM was downloaded from The Cancer Genome Atlas (TCGA) database and randomly divided into training group and test group. Survival statistical analysis and machine-learning approaches were performed on the RNA sequencing data of CM to develop a prognostic signature. Using univariable Cox proportional hazards regression, random survival forest algorithm, and receiver operating characteristic (ROC) in the training group, the four-mRNA signature including CD276, UQCRFS1, HAPLN3, and PIP4P1 was screened out. The four-mRNA signature could divide patients into low-risk and high-risk groups with different survival outcomes (log-rank p < 0.001). The predictive efficacy of the four-mRNA signature was confirmed in the test group, the whole TCGA group, and the independent GSE65904 (log-rank p < 0.05). The independence of the four-mRNA signature in prognostic prediction was demonstrated by multivariate Cox analysis. ROC and timeROC analyses showed that the efficiency of the signature in survival prediction was better than other clinical variables such as melanoma Clark level and tumor stage. This study highlights that the four-mRNA model could be used as a prognostic signature for CM patients with potential clinical application value.
Keywords: MRNA expression data; cutaneous melanoma; machine learning; prognostic signature; random survival forest.
Copyright © 2021 Bai, Wang, Liu and Lu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
LinkOut - more resources
Full Text Sources
Research Materials

