N-terminus-independent activation of c-Src via binding to a tetraspan(in) TM4SF5 in hepatocellular carcinoma is abolished by the TM4SF5 C-terminal peptide application
- PMID: 34335982
- PMCID: PMC8315060
- DOI: 10.7150/thno.58739
N-terminus-independent activation of c-Src via binding to a tetraspan(in) TM4SF5 in hepatocellular carcinoma is abolished by the TM4SF5 C-terminal peptide application
Abstract
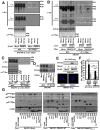
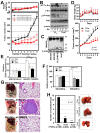
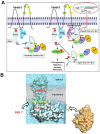
Active c-Src non-receptor tyrosine kinase localizes to the plasma membrane via N-terminal lipid modification. Membranous c-Src causes cancer initiation and progression. Even though transmembrane 4 L six family member 5 (TM4SF5), a tetraspan(in), can be involved in this mechanism, the molecular and structural influence of TM4SF5 on c-Src remains unknown. Methods: Here, we investigated molecular and structural details by which TM4SF5 regulated c-Src devoid of its N-terminus and how cell-penetrating peptides were able to interrupt c-Src activation via interference of c-Src-TM4SF5 interaction in hepatocellular carcinoma models. Results: The TM4SF5 C-terminus efficiently bound the c-Src SH1 kinase domain, efficiently to the inactively-closed form. The complex involved protein tyrosine phosphatase 1B able to dephosphorylate Tyr530. The c-Src SH1 domain alone, even in a closed form, bound TM4SF5 to cause c-Src Tyr419 and FAK Y861 phosphorylation. Homology modeling and molecular dynamics simulation studies predicted the directly interfacing residues, which were further validated by mutational studies. Cell penetration of TM4SF5 C-terminal peptides blocked the interaction of TM4SF5 with c-Src and prevented c-Src-dependent tumor initiation and progression in vivo. Conclusions: Collectively, these data demonstrate that binding of the TM4SF5 C-terminus to the kinase domain of inactive c-Src leads to its activation. Because this binding can be abolished by cell-penetrating peptides containing the TM4SF5 C-terminus, targeting this direct interaction may be an effective strategy for developing therapeutics that block the development and progression of hepatocellular carcinoma.
Keywords: PTPIB; TM4SF5; c-Src; metastasis; protein-protein interaction.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures





Similar articles
-
The COOH-terminus of TM4SF5 in hepatoma cell lines regulates c-Src to form invasive protrusions via EGFR Tyr845 phosphorylation.Biochim Biophys Acta. 2013 Mar;1833(3):629-42. doi: 10.1016/j.bbamcr.2012.11.026. Epub 2012 Dec 6. Biochim Biophys Acta. 2013. PMID: 23220047
-
Tetraspan TM4SF5-dependent direct activation of FAK and metastatic potential of hepatocarcinoma cells.J Cell Sci. 2012 Dec 15;125(Pt 24):5960-73. doi: 10.1242/jcs.100586. Epub 2012 Oct 17. J Cell Sci. 2012. PMID: 23077174 Free PMC article.
-
CD133-induced TM4SF5 expression promotes sphere growth via recruitment and blocking of protein tyrosine phosphatase receptor type F (PTPRF).Cancer Lett. 2018 Dec 1;438:219-231. doi: 10.1016/j.canlet.2018.09.009. Epub 2018 Sep 11. Cancer Lett. 2018. PMID: 30217560
-
TM4SF5-mediated protein-protein networks and tumorigenic roles.BMB Rep. 2014 Sep;47(9):483-7. doi: 10.5483/bmbrep.2014.47.9.146. BMB Rep. 2014. PMID: 25027595 Free PMC article. Review.
-
TM4SF5-Mediated Regulation of Hepatocyte Transporters during Metabolic Liver Diseases.Int J Mol Sci. 2022 Jul 29;23(15):8387. doi: 10.3390/ijms23158387. Int J Mol Sci. 2022. PMID: 35955521 Free PMC article. Review.
Cited by
-
Liver-originated small extracellular vesicles with TM4SF5 target brown adipose tissue for homeostatic glucose clearance.J Extracell Vesicles. 2022 Sep;11(9):e12262. doi: 10.1002/jev2.12262. J Extracell Vesicles. 2022. PMID: 36063136 Free PMC article.
-
Isoxazole-based molecules restore NK cell immune surveillance in hepatocarcinogenesis by targeting TM4SF5 and SLAMF7 linkage.Signal Transduct Target Ther. 2025 Jan 20;10(1):15. doi: 10.1038/s41392-024-02106-6. Signal Transduct Target Ther. 2025. PMID: 39828766 Free PMC article.
-
Glucose-mediated mitochondrial reprogramming by cholesterol export at TM4SF5-enriched mitochondria-lysosome contact sites.Cancer Commun (Lond). 2024 Jan;44(1):47-75. doi: 10.1002/cac2.12510. Epub 2023 Dec 22. Cancer Commun (Lond). 2024. PMID: 38133457 Free PMC article.
-
Systemic TM4SF5 overexpression in ApcMin/+ mice promotes hepatic portal hypertension associated with fibrosis.BMB Rep. 2022 Dec;55(12):609-614. doi: 10.5483/BMBRep.2022.55.12.104. BMB Rep. 2022. PMID: 36104259 Free PMC article.
-
Role of c-Src in Carcinogenesis and Drug Resistance.Cancers (Basel). 2023 Dec 20;16(1):32. doi: 10.3390/cancers16010032. Cancers (Basel). 2023. PMID: 38201459 Free PMC article. Review.
References
-
- Yeatman TJ. A renaissance for SRC. Nat Rev Cancer. 2004;4:470–80. - PubMed
-
- Roskoski R Jr. Src protein-tyrosine kinase structure, mechanism, and small molecule inhibitors. Pharmacol Res. 2015;94:9–25. - PubMed
-
- Gargalionis AN, Karamouzis MV, Papavassiliou AG. The molecular rationale of Src inhibition in colorectal carcinomas. Intern J Cancer. 2014;134:2019–29. - PubMed
-
- Roskoski R Jr. Src protein-tyrosine kinase structure and regulation. Biochem Biophys Res Commun. 2004;324:1155–64. - PubMed
-
- Moarefi I, LaFevre-Bernt M, Sicheri F, Huse M, Lee CH, Kuriyan J. et al. Activation of the Src-family tyrosine kinase Hck by SH3 domain displacement. Nature. 1997;385:650–3. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

