Biosynthetic Cyclization Catalysts for the Assembly of Peptide and Polyketide Natural Products
- PMID: 34335987
- PMCID: PMC8320681
- DOI: 10.1002/cctc.202001886
Biosynthetic Cyclization Catalysts for the Assembly of Peptide and Polyketide Natural Products
Abstract
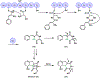
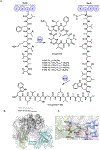
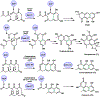
Many biologically active natural products are synthesized by nonribosomal peptide synthetases (NRPSs), polyketide synthases (PKSs) and their hybrids. These megasynthetases contain modules possessing distinct catalytic domains that allow for substrate initiation, chain extension, processing and termination. At the end of a module, a terminal domain, usually a thioesterase (TE), is responsible for catalyzing the release of the NRPS or PKS as a linear or cyclized product. In this review, we address the general cyclization mechanism of the TE domain, including oligomerization and the fungal C-C bond forming Claisen-like cyclases (CLCs). Additionally, we include examples of cyclization catalysts acting within or at the end of a module. Furthermore, condensation-like (CT) domains, terminal reductase (R) domains, reductase-like domains that catalyze Dieckmann condensation (RD), thioesterase-like Dieckmann cyclases, trans-acting TEs from the penicillin binding protein (PBP) enzyme family, product template (PT) domains and others will also be reviewed. The studies summarized here highlight the remarkable diversity of NRPS and PKS cyclization catalysts for the production of biologically relevant, complex cyclic natural products and related compounds.
Keywords: Biocatalysis; Enzymes; Natural Products; Nonribosomal Peptide Synthetase; Polyketide Synthase.
Figures
















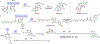


References
-
- Du L, Lou L, Nat. Prod. Rep 2010, 27, 255–278. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
