Metabolic Profile and Neurogenic Potential of Human Amniotic Fluid Stem Cells From Normal vs. Fetus-Affected Gestations
- PMID: 34336852
- PMCID: PMC8322743
- DOI: 10.3389/fcell.2021.700634
Metabolic Profile and Neurogenic Potential of Human Amniotic Fluid Stem Cells From Normal vs. Fetus-Affected Gestations
Abstract
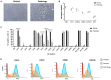
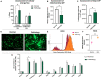
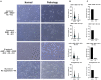
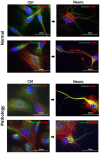
Human amniotic fluid stem cells (hAFSCs) possess some characteristics with mesenchymal stem cells (MSCs) and embryonic stem cells and have a broader differentiation potential compared to MSCs derived from other sources. Although hAFSCs are widely researched, their analysis mainly involves stem cells (SCs) obtained from normal, fetus-unaffected gestations. However, in clinical settings, knowledge about hAFSCs from normal gestations could be poorly translational, as hAFSCs from healthy and fetus-diseased gestations may differ in their differentiation and metabolic potential. Therefore, a more thorough investigation of hAFSCs derived from pathological gestations would provide researchers with the knowledge about the general characteristics of these cells that could be valuable for further scientific investigations and possible future clinical applicability. The goal of this study was to look into the neurogenic and metabolic potential of hAFSCs derived from diseased fetuses, when gestations were concomitant with polyhydramnios and compare them to hAFSCs derived from normal fetuses. Results demonstrated that these cells are similar in gene expression levels of stemness markers (SOX2, NANOG, LIN28A, etc.). However, they differ in expression of CD13, CD73, CD90, and CD105, as flow cytometry analysis revealed higher expression in hAFSCs from unaffected gestations. Furthermore, hAFSCs from "Normal" and "Pathology" groups were different in oxidative phosphorylation rate, as well as level of ATP and reactive oxygen species production. Although the secretion of neurotrophic factors BDNF and VEGF was of comparable degree, as evaluated with enzyme-linked immunosorbent assay (ELISA) test, hAFSCs from normal gestations were found to be more prone to neurogenic differentiation, compared to hAFSCs from polyhydramnios. Furthermore, hAFSCs from polyhydramnios were distinguished by higher secretion of pro-inflammatory cytokine TNFα, which was significantly downregulated in differentiated cells. Overall, these observations show that hAFSCs from pathological gestations with polyhydramnios differ in metabolic and inflammatory status and also possess lower neurogenic potential compared to hAFSCs from normal gestations. Therefore, further in vitro and in vivo studies are necessary to dissect the potential of hAFSCs from polyhydramnios in stem cell-based therapies. Future studies should also search for strategies that could improve the characteristics of hAFSCs derived from diseased fetuses in order for those cells to be successfully applied for regenerative medicine purposes.
Keywords: cell differentiation; energy metabolism; mesenchymal stem cells; neurogenesis; polyhydramnios.
Copyright © 2021 Valiulienė, Zentelytė, Beržanskytė and Navakauskienė.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

