Critical Presentation of a Severe Acute Respiratory Syndrome Coronavirus 2 Reinfection: A Case Report
- PMID: 34337095
- PMCID: PMC8320276
- DOI: 10.1093/ofid/ofab329
Critical Presentation of a Severe Acute Respiratory Syndrome Coronavirus 2 Reinfection: A Case Report
Abstract
Background: Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) reinfections have been reported; however, most cases are milder than the primary infection. We report the first case of a life-threatening critical presentation of a SARS-CoV-2 reinfection.
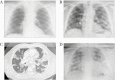
Methods: A 62-year-old man from Palamós (Spain) suffered a first mild coronavirus disease 2019 (COVID-19) episode in March 2020, confirmed by 2 independent SARS-CoV-2 nasopharyngeal polymerase chain reaction (PCR) assays and a normal radiograph. He recovered completely and tested negative on 2 consecutive PCRs. In August 2020, the patient developed a second SARS-CoV-2 infection with life-threatening bilateral pneumonia and Acute respiratory distress syndrome criteria, requiring COVID-19-specific treatment (remdesivir + dexamethasone) plus high-flow oxygen therapy. Nasopharyngeal swabs from the second episode were obtained for virus quantification by real-time PCR, for virus outgrowth and sequencing. In addition, plasma and peripheral blood mononuclear cells during the hospitalization period were used to determine SARS-CoV-2-specific humoral and T-cell responses.
Results: Genomic analysis of SARS-CoV-2 showed that the virus had probably originated shortly before symptom onset. When the reinfection occurred, the subject showed a weak immune response, with marginal humoral and specific T-cell responses against SARS-CoV-2. All antibody isotypes tested as well as SARS-CoV-2 neutralizing antibodies increased sharply after day 8 postsymptoms. A slight increase of T-cell responses was observed at day 19 after symptom onset.
Conclusions: The reinfection was firmly documented and occurred in the absence of robust preexisting humoral and cellular immunity. SARS-CoV-2 immunity in some subjects is unprotective and/or short-lived; therefore, SARS-CoV-2 vaccine schedules inducing long-term immunity will be required to bring the pandemic under control.
Keywords: 19; 2; CoV; SARS; immune responses; life; reinfection; secondary infection; threatening COVID.
© The Author(s) 2021. Published by Oxford University Press on behalf of Infectious Diseases Society of America.
Figures

References
-
- Prado-Vivar B, Becerra-Wong M, Guadalupe JJ, et al. COVID-19 re-infection by a phylogenetically distinct SARS-CoV-2 variant, first confirmed event in South America. SSRN [Preprint]. Posted online 8 September 2020. doi:10.2139/ssrn/3686174.
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

