PScL-HDeep: image-based prediction of protein subcellular location in human tissue using ensemble learning of handcrafted and deep learned features with two-layer feature selection
- PMID: 34337652
- PMCID: PMC8574991
- DOI: 10.1093/bib/bbab278
PScL-HDeep: image-based prediction of protein subcellular location in human tissue using ensemble learning of handcrafted and deep learned features with two-layer feature selection
Abstract
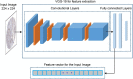
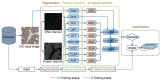
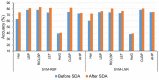
Protein subcellular localization plays a crucial role in characterizing the function of proteins and understanding various cellular processes. Therefore, accurate identification of protein subcellular location is an important yet challenging task. Numerous computational methods have been proposed to predict the subcellular location of proteins. However, most existing methods have limited capability in terms of the overall accuracy, time consumption and generalization power. To address these problems, in this study, we developed a novel computational approach based on human protein atlas (HPA) data, referred to as PScL-HDeep, for accurate and efficient image-based prediction of protein subcellular location in human tissues. We extracted different handcrafted and deep learned (by employing pretrained deep learning model) features from different viewpoints of the image. The step-wise discriminant analysis (SDA) algorithm was applied to generate the optimal feature set from each original raw feature set. To further obtain a more informative feature subset, support vector machine-based recursive feature elimination with correlation bias reduction (SVM-RFE + CBR) feature selection algorithm was applied to the integrated feature set. Finally, the classification models, namely support vector machine with radial basis function (SVM-RBF) and support vector machine with linear kernel (SVM-LNR), were learned on the final selected feature set. To evaluate the performance of the proposed method, a new gold standard benchmark training dataset was constructed from the HPA databank. PScL-HDeep achieved the maximum performance on 10-fold cross validation test on this dataset and showed a better efficacy over existing predictors. Furthermore, we also illustrated the generalization ability of the proposed method by conducting a stringent independent validation test.
Keywords: bioimage analysis; deep learned features; feature selection; handcrafted features; protein subcellular location.
© The Author(s) 2021. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures





References
-
- Yang F, Xu Y-Y, Wang S-T, et al. . Image-based classification of protein subcellular location patterns in human reproductive tissue by ensemble learning global and local features. Neurocomputing 2014;131:113–23.
-
- Hung M-C, Link W. Protein localization in disease and therapy. J Cell Sci 2011;124:3381–92. - PubMed
-
- Kajiwara D, Minamiguchi K, Seki M, et al. . Effect of a new type androgen receptor antagonist, TAS3681, on ligand-independent AR activation through its AR downregulation activity. J Clin Oncol 2016;34:199–9. - PubMed
-
- Thul PJ, Akesson L, Wiking M, et al. . A subcellular map of the human proteome. Science 2017;356:eaal3321. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

