Method for fast lipid reconstruction and removal processing in 1 H MRSI of the brain
- PMID: 34337788
- PMCID: PMC8568649
- DOI: 10.1002/mrm.28949
Method for fast lipid reconstruction and removal processing in 1 H MRSI of the brain
Abstract
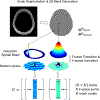
Purpose: To develop a new rapid spatial filtering method for lipid removal, fast lipid reconstruction and removal processing (FLIP), which selectively isolates and removes interfering lipid signals from outside the brain in a full-FOV 2D MRSI and whole-brain 3D echo planar spectroscopic imaging (EPSI).
Theory and methods: FLIP uses regularized least-squares regression based on spatial prior information from MRI to selectively remove lipid signals originating from the scalp and measure the brain metabolite signals with minimum cross contamination. FLIP is a noniterative approach, thus allowing a rapid processing speed, and uses only spatial information without any spectral priors. The performance of FLIP was compared with the Papoulis-Gerchberg algorithm (PGA), Hankel singular value decomposition (HSVD), and fast image reconstruction with L2 regularization (L2).
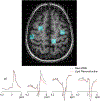
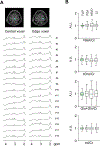
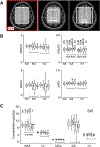
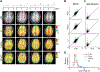
Results: FLIP in both 2D and 3D MRSI resulted in consistent metabolite quantification in a greater number of voxels with less concentration variation than other algorithms, demonstrating effective and robust lipid-removal performance. The percentage of voxels that met quality criteria with FLIP, PGA, HSVD, and L2 processing were 90%, 57%, 29%, and 42% in 2D MRSI, and 80%, 75%, 76%, and 74% in 3D EPSI, respectively. The quantification results of full-FOV MRSI using FLIP were comparable to those of volume-localized MRSI, while allowing significantly increased spatial coverage. FLIP performed the fastest in 2D MRSI.
Conclusion: FLIP is a new lipid-removal algorithm that promises fast and effective lipid removal with improved volume coverage in MRSI.
Keywords: full FOV MRSI; lipid reconstruction; lipid removal; spatial-domain post processing; whole brain MRSI.
© 2021 International Society for Magnetic Resonance in Medicine.
Figures
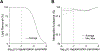

References
-
- Pijnappel WWF, Vandenboogaart A, Debeer R, Vanormondt D. SVD-Based Quantification of Magnetic-Resonance Signals. Journal of Magnetic Resonance. 1992;97(1):122–134.
-
- Barkhuijsen H, Debeer R, Vanormondt D. Improved Algorithm for Noniterative Time-Domain Model-Fitting to Exponentially Damped Magnetic-Resonance Signals. Journal of Magnetic Resonance. 1987;73(3):553–557.
-
- Kumaresan R, Tufts DW. Estimating the Parameters of Exponentially Damped Sinusoids and Pole-Zero Modeling in Noise. Ieee Transactions on Acoustics Speech and Signal Processing. 1982;30(6):833–840.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

