High-Throughput Crystallography Reveals Boron-Containing Inhibitors of a Penicillin-Binding Protein with Di- and Tricovalent Binding Modes
- PMID: 34337941
- PMCID: PMC9282634
- DOI: 10.1021/acs.jmedchem.1c00717
High-Throughput Crystallography Reveals Boron-Containing Inhibitors of a Penicillin-Binding Protein with Di- and Tricovalent Binding Modes
Abstract
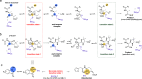
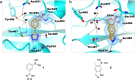
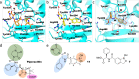
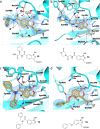
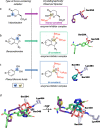
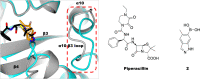
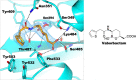
The effectiveness of β-lactam antibiotics is increasingly compromised by β-lactamases. Boron-containing inhibitors are potent serine-β-lactamase inhibitors, but the interactions of boron-based compounds with the penicillin-binding protein (PBP) β-lactam targets have not been extensively studied. We used high-throughput X-ray crystallography to explore reactions of a boron-containing fragment set with the Pseudomonas aeruginosa PBP3 (PaPBP3). Multiple crystal structures reveal that boronic acids react with PBPs to give tricovalently linked complexes bonded to Ser294, Ser349, and Lys484 of PaPBP3; benzoxaboroles react with PaPBP3 via reaction with two nucleophilic serines (Ser294 and Ser349) to give dicovalently linked complexes; and vaborbactam reacts to give a monocovalently linked complex. Modifications of the benzoxaborole scaffold resulted in a moderately potent inhibition of PaPBP3, though no antibacterial activity was observed. Overall, the results further evidence the potential for the development of new classes of boron-based antibiotics, which are not compromised by β-lactamase-driven resistance.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


References
-
- Adam M.; Fraipont C.; Rhazi N.; Nguyen-Distèche M.; Lakaye B.; Frère J. M.; Devreese B.; Van Beeumen J.; van Heijenoort Y.; van Heijenoort J.; Ghuysen J. M. The Bimodular G57-V577 Polypeptide Chain of the Class B Penicillin-Binding Protein 3 of Escherichia Coli Catalyzes Peptide Bond Formation from Thiolesters and Does Not Catalyze Glycan Chain Polymerization from the Lipid II Intermediate. J. Bacteriol. 1997, 179, 6005–6009. 10.1128/jb.179.19.6005-6009.1997. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

