Exendin-4 stimulates autophagy in pancreatic β-cells via the RAPGEF/EPAC-Ca2+-PPP3/calcineurin-TFEB axis
- PMID: 34338148
- PMCID: PMC9037459
- DOI: 10.1080/15548627.2021.1956123
Exendin-4 stimulates autophagy in pancreatic β-cells via the RAPGEF/EPAC-Ca2+-PPP3/calcineurin-TFEB axis
Abstract
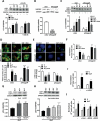
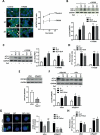
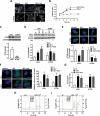
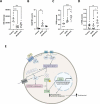
Macroautophagy/autophagy is critical for the regulation of pancreatic β-cell mass and its deregulation has been implicated in the pathogenesis of type 2 diabetes (T2D). We have previously shown that treatment of pancreatic β-cells with the GLP1R (glucagon like peptide 1 receptor) agonist exendin-4 stimulates autophagic flux in a setting of chronic nutrient excess. The aim of this study was to identify the underlying pathways contributing to enhanced autophagic flux.Pancreatic β-cells (INS-1E),mouse and human islets were treated with glucolipotoxic stress (0.5 mM palmitate and 25 mM glucose) in the presence of exendin-4. Consistent with our previous work, exendin-4 stimulated autophagic flux. Using chemical inhibitors and siRNA knockdown, we identified RAPGEF4/EPAC2 (Rap guanine nucleotide exchange factor 4) and downstream calcium signaling to be essential for regulation of autophagic flux by exendin-4. This pathway was independent of AMPK and MTOR signaling. Further analysis identified PPP3/calcineurin and its downstream regulator TFEB (transcription factor EB) as key proteins mediating exendin-4 induced autophagy. Importantly, inhibition of this pathway prevented exendin-4-mediated cell survival and overexpression of TFEB mimicked the cell protective effects of exendin-4 in INS-1E and human islets. Moreover, treatment of db/db mice with exendin-4 for 21 days increased the expression of lysosomal markers within the pancreatic islets. Collectively our data identify the RAPGEF4/EPAC2-calcium-PPP3/calcineurin-TFEB axis as a key mediator of autophagic flux, lysosomal function and cell survival in pancreatic β-cells. Pharmacological modulation of this axis may offer a novel therapeutic target for the treatment of T2D.Abbreviations: AKT1/protein kinase B: AKT serine/threonine kinase 1; AMPK: 5' AMP-activated protein kinase; CAMKK: calcium/calmodulin-dependent protein kinase kinase; cAMP: cyclic adenosine monophosphate; CASP3: caspase 3; CREB: cAMP response element-binding protein; CTSD: cathepsin D; Ex4: exendin-4(1-39); GLP-1: glucagon like peptide 1; GLP1R: glucagon like peptide 1 receptor; GLT: glucolipotoxicity; INS: insulin; MTOR: mechanistic target of rapamycin kinase; NFAT: nuclear factor of activated T-cells; PPP3/calcineurin: protein phosphatase 3; PRKA/PKA: protein kinase cAMP activated; RAPGEF3/EPAC1: Rap guanine nucleotide exchange factor 3; RAPGEF4/EPAC2: Rap guanine nucleotide exchange factor 4; SQSTM1/p62: sequestosome 1; T2D: type 2 diabetes; TFEB: transcription factor EB.
Keywords: Autophagy; GLP1R agonists; PPP3/calcineurin; RAPGEF4; TFEB; diabetes; pancreatic β-cell.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures




References
-
- Kaur J, Debnath J. Autophagy at the crossroads of catabolism and anabolism. Nat Rev Mol Cell Biol. 2015;16(8):461–472. - PubMed
-
- Lavallard VJ, Meijer AJ, Codogno P, et al. Autophagy, signalling and obesity. Pharmacol Res. 2012;66:513–525. - PubMed
-
- Kim KH, Lee MS. Autophagy–a key player in cellular and body metabolism. Nat Rev Endocrinol. 2014;10(6):322–337. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
