Uniparental nuclear inheritance following bisexual mating in fungi
- PMID: 34338631
- PMCID: PMC8412948
- DOI: 10.7554/eLife.66234
Uniparental nuclear inheritance following bisexual mating in fungi
Abstract
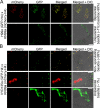
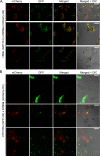
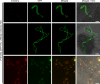
Some remarkable animal species require an opposite-sex partner for their sexual development but discard the partner's genome before gamete formation, generating hemi-clonal progeny in a process called hybridogenesis. Here, we discovered a similar phenomenon, termed pseudosexual reproduction, in a basidiomycete human fungal pathogen, Cryptococcus neoformans, where exclusive uniparental inheritance of nuclear genetic material was observed during bisexual reproduction. Analysis of strains expressing fluorescent reporter proteins revealed instances where only one of the parental nuclei was present in the terminal sporulating basidium. Whole-genome sequencing revealed that the nuclear genome of the progeny was identical with one or the other parental genome. Pseudosexual reproduction was also detected in natural isolate crosses where it resulted in mainly MATα progeny, a bias observed in Cryptococcus ecological distribution as well. The mitochondria in these progeny were inherited from the MATa parent, resulting in nuclear-mitochondrial genome exchange. The meiotic recombinase Dmc1 was found to be critical for pseudosexual reproduction. These findings reveal a novel, and potentially ecologically significant, mode of eukaryotic microbial reproduction that shares features with hybridogenesis in animals.
Keywords: Cryptococcus neoformans; cryptococcus neoformans; genetics; genomics; hybridogenesis; infectious disease; meiosis; microbiology; nuclear migration; pseudosexual reproduction; sexual parasitism.
Plain language summary
Sexual reproduction enables organisms to recombine their genes to generate progeny that have higher levels of evolutionary fitness. This process requires reproductive cells – like the sperm and egg – to fuse together and mix their two genomes, resulting in offspring that are genetically distinct from their parents. In a disease-causing fungus called Cryptococcus neoformans, sexual reproduction occurs when two compatible mating types (MATa and MATα) merge together to form long branched filaments called hyphae. Cells in the hyphae contain two nuclei – one from each parent – which fuse in specialized cells at the end of the branches called basidia. The fused nucleus is then divided into four daughter nuclei, which generate spores that can develop into new organisms. In nature, the mating types of C. neoformans exhibit a peculiar distribution where MATα represents 95% or more of the population. However, it is not clear how this fungus successfully reproduces with such an unusually skewed distribution of mating types. To investigate this further, Yadav et al. tracked the reproductive cycle of C. neoformans applying genetic techniques, fluorescence microscopy, and whole-genome sequencing. This revealed that during hyphal branching some cells lose the nucleus of one of the two mating types. As a result, the nuclei of the generated spores only contain genetic information from one parent. Yadav et al. named this process pseudosexual reproduction as it defies the central benefit of sex, which is to produce offspring with a new combination of genetic information. Further experiments showed that this unconventional mode of reproduction can be conducted by fungi isolated from both environmental samples and clinical patient samples. This suggests that pseudosexual reproduction is a widespread and conserved process that may provide significant evolutionary benefits. C. neoformans represents a flexible and adaptable model organism to explore the impact and evolutionary advantages of sex. Further studies of the unique reproductive strategies employed by this fungus may improve the understanding of similar processes in other eukaryotes, including animals and plants. This research may also have important implications for understanding and controlling the growth of other disease-causing microbes.
© 2021, Yadav et al.
Conflict of interest statement
VY, SS, JH No competing interests declared
Figures










Similar articles
-
Obligate sexual reproduction of a homothallic fungus closely related to the Cryptococcus pathogenic species complex.Elife. 2022 Jun 17;11:e79114. doi: 10.7554/eLife.79114. Elife. 2022. PMID: 35713948 Free PMC article.
-
The Pheromone and Pheromone Receptor Mating-Type Locus Is Involved in Controlling Uniparental Mitochondrial Inheritance in Cryptococcus.Genetics. 2020 Mar;214(3):703-717. doi: 10.1534/genetics.119.302824. Epub 2019 Dec 30. Genetics. 2020. PMID: 31888949 Free PMC article.
-
The mating type-specific homeodomain genes SXI1 alpha and SXI2a coordinately control uniparental mitochondrial inheritance in Cryptococcus neoformans.Curr Genet. 2007 Mar;51(3):187-95. doi: 10.1007/s00294-006-0115-9. Curr Genet. 2007. PMID: 17186242
-
[Mating types, sexual reproduction and ploidy in fungi: effects on virulence].Mikrobiyol Bul. 2009 Jul;43(3):507-13. Mikrobiyol Bul. 2009. PMID: 19795629 Review. Turkish.
-
Unisexual versus bisexual mating in Cryptococcus neoformans: Consequences and biological impacts.Fungal Genet Biol. 2015 May;78:65-75. doi: 10.1016/j.fgb.2014.08.008. Epub 2014 Aug 27. Fungal Genet Biol. 2015. PMID: 25173822 Free PMC article. Review.
Cited by
-
The Fungal Kingdom as a Rosetta Stone for biological discovery.Curr Biol. 2025 Jun 9;35(11):R427-R433. doi: 10.1016/j.cub.2025.04.013. Curr Biol. 2025. PMID: 40494291 Free PMC article.
-
A conceptual framework for nomenclatural stability and validity of medically important fungi: a proposed global consensus guideline for fungal name changes supported by ABP, ASM, CLSI, ECMM, ESCMID-EFISG, EUCAST-AFST, FDLC, IDSA, ISHAM, MMSA, and MSGERC.J Clin Microbiol. 2023 Nov 21;61(11):e0087323. doi: 10.1128/jcm.00873-23. Epub 2023 Oct 26. J Clin Microbiol. 2023. PMID: 37882528 Free PMC article.
-
Invasive Californian death caps develop mushrooms unisexually and bisexually.Nat Commun. 2023 Oct 24;14(1):6560. doi: 10.1038/s41467-023-42317-z. Nat Commun. 2023. PMID: 37875491 Free PMC article.
-
Tracing the evolution and genomic dynamics of mating-type loci in Cryptococcus pathogens and closely related species.bioRxiv [Preprint]. 2025 Feb 16:2025.02.12.637874. doi: 10.1101/2025.02.12.637874. bioRxiv. 2025. PMID: 39990455 Free PMC article. Preprint.
-
Insights into Fungal Mitochondrial Genomes and Inheritance Based on Current Findings from Yeast-like Fungi.J Fungi (Basel). 2024 Jun 21;10(7):441. doi: 10.3390/jof10070441. J Fungi (Basel). 2024. PMID: 39057326 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

