The C-terminal region of yeast ubiquitin-protein ligase Not4 mediates its cellular localization and stress response
- PMID: 34338747
- PMCID: PMC8370887
- DOI: 10.1093/femsle/fnab097
The C-terminal region of yeast ubiquitin-protein ligase Not4 mediates its cellular localization and stress response
Erratum in
-
Corrigendum to: The C-terminal region of yeast ubiquitin-protein ligase Not4 mediates its cellular localization and stress response.FEMS Microbiol Lett. 2021 Oct 9;368(18):fnab122. doi: 10.1093/femsle/fnab122. FEMS Microbiol Lett. 2021. PMID: 34628507 Free PMC article. No abstract available.
Abstract
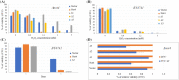
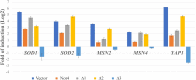
Transient modification of the environment involves the expression of specific genes and degradation of mRNAs and proteins. How these events are linked is poorly understood. CCR4-NOT is an evolutionary conserved complex involved in transcription initiation and mRNA degradation. In this paper, we report that the yeast Not4 localizes in cytoplasmic foci after cellular stress. We focused our attention on the functional characterization of the C-terminus of the Not4 protein. Molecular dissection of this region indicates that the removal of the last 120 amino acids, does not affect protein localization and function, in that the protein is still able to suppress the thermosensitivity observed in the not4Δ mutant. In addition, such shortened form of Not4, as well its absence, increases the transcription of stress-responsive genes conferring to the cell high resistance to the oxidative stress. On the contrary, the last C-terminal 211 amino acids are required for proper Not4 localization at cytoplasmic foci after stress. This truncated version of Not4 fails to increase the transcription of the stress genes, is more stable and seems to be toxic to cells undergoing oxidative stress.
Keywords: E3 ubiquitin ligase; gene expression; protein aggregation; stress response; yeast.
© The Author(s) 2021. Published by Oxford University Press on behalf of FEMS.
Figures


References
-
- Bhaskar V, Roudko V, Basquin Jet al. Structure and RNA-binding properties of the Not1-Not2-Not5 module of the yeast Ccr4-Not complex. Nat Struct Mol Biol. 2013;20:1281–8. - PubMed
-
- Brachmann CB, Davies A, Cost GJet al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–32. - PubMed
-
- Burkhard P, Stetefeld J, Strelkov SV. Coiled coils: a highly versatile protein folding motif. Trends Cell Biol. 2001;11:82–8. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

