Design and Evaluation of Multiplex One-Step Reverse Transcription PCR-Dipstick Chromatography Method for the Analysis of Seven Respiratory Pathogens
- PMID: 34338833
- PMCID: PMC8326646
- DOI: 10.1007/s00284-021-02621-7
Design and Evaluation of Multiplex One-Step Reverse Transcription PCR-Dipstick Chromatography Method for the Analysis of Seven Respiratory Pathogens
Abstract
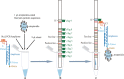
Influenza A, influenza B, severe acute respiratory syndrome coronavirus 2, adenovirus, respiratory syncytial virus, Mycoplasma pneumoniae, and Chlamydophila pneumoniae are common pathogens that can cause severe pneumonia and other symptoms, resulting in acute lower respiratory tract infections. The objective of this study was to design and evaluate a sensitive and specific multiplex one-step reverse transcription PCR (RT-PCR)-dipstick chromatography method for simultaneous rapid detection of these seven pathogens. Streptavidin-coated blue latex particles were used to read out a positive signal. Based on the DNA-DNA hybridization of oligonucleotide sequences (Tag) for forward primer with the complementary oligonucleotide sequence (cTag) on the dipstick and biotin-streptavidin interactions, PCR products were able to be illuminated visually on the dipstick. The specificity and the limit of detection (LOD) were also evaluated. Moreover, the clinical performance of this method was compared with Sanger sequencing for 896 samples. No cross reaction with other pathogens was found, confirming the high specificity of this method. The LOD was 10 copies/µL for each of the tested pathogens, and the whole procedure took less than 40 min. Using 896 samples, the sensitivity and specificity were shown to be no lower than 94.5%. The positive predictive value was higher than 82.1%, and the negative predictive value was higher than 99.5%. The kappa value between the PCR-dipstick chromatography method and Sanger sequencing ranged from 0.869 to 0.940. In summary, our one-step RT-PCR-dipstick chromatography method is a sensitive and specific tool for rapidly detecting multiplex respiratory pathogens.
© 2021. The Author(s), under exclusive licence to Springer Science+Business Media, LLC, part of Springer Nature.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures




References
-
- Nair H, Simoes EA, Rudan I, Gessner BD, Azziz-Baumgartner E, Zhang J, Feikin DR, Mackenzie GA, Moiisi JC, Roca A, Baggett HC, Zaman SM, Singleton RJ, Lucero MG, Chandran A, Gentile A, Cohen C, Krishnan A, Bhutta ZA, Arguedas A, Clara AW, Andrade AL, Ope M, Ruvinsky RO, Hortal M, Mccracken JP, Madhi SA, Bruce N, Qazi SA, Morris SS, El AS, Weber MW, Scott J, Brooks WA, Breiman RF, Campbell H. Global and regional burden of hospital admissions for severe acute lower respiratory infections in young children in 2010: a systematic analysis. Lancet. 2013;381(9875):1380–1390. doi: 10.1016/S0140-6736(12)61901-1. - DOI - PMC - PubMed
-
- Collaborators GDAI. Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990–2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet. 2016;388(10053):1545–1602. doi: 10.1016/S0140-6736(16)31678-6. - DOI - PMC - PubMed
-
- Jain S, Williams DJ, Arnold SR, Ampofo K, Bramley AM, Reed C, Stockmann C, Anderson EJ, Grijalva CG, Self WH, Zhu Y, Patel A, Hymas W, Chappell JD, Kaufman RA, Kan JH, Dansie D, Lenny N, Hillyard DR, Haynes LM, Levine M, Lindstrom S, Winchell JM, Katz JM, Erdman D, Schneider E, Hicks LA, Wunderink RG, Edwards KM, Pavia AT, Mccullers JA, Finelli L. Community-acquired pneumonia requiring hospitalization among U.S. Children. New Engl J Med. 2015;372(9):835–845. doi: 10.1056/NEJMoa1405870. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

