Identifying Small Open Reading Frames in Prokaryotes with Ribosome Profiling
- PMID: 34339296
- PMCID: PMC8765392
- DOI: 10.1128/JB.00294-21
Identifying Small Open Reading Frames in Prokaryotes with Ribosome Profiling
Abstract
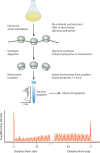
Small proteins encoded by open reading frames (ORFs) shorter than 50 codons (small ORFs [sORFs]) are often overlooked by annotation engines and are difficult to characterize using traditional biochemical techniques. Ribosome profiling has tremendous potential to empirically improve the annotations of prokaryotic genomes. Recent improvements in ribosome profiling methods for bacterial model organisms have revealed many new sORFs in well-characterized genomes. Antibiotics that trap ribosomes just after initiation have played a key role in these developments by allowing the unambiguous identification of the start codons (and, hence, the reading frame) for novel ORFs. Here, we describe these new methods and highlight critical controls and considerations for adapting ribosome profiling to different prokaryotic species.
Keywords: genome annotation; ribosome profiling; sORF; small protein.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

