The pentose phosphate pathway constitutes a major metabolic hub in pathogenic Francisella
- PMID: 34339477
- PMCID: PMC8360588
- DOI: 10.1371/journal.ppat.1009326
The pentose phosphate pathway constitutes a major metabolic hub in pathogenic Francisella
Abstract
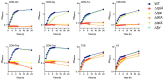
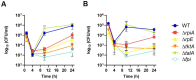
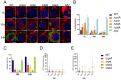
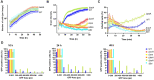
Metabolic pathways are now considered as intrinsic virulence attributes of pathogenic bacteria and thus represent potential targets for antibacterial strategies. Here we focused on the role of the pentose phosphate pathway (PPP) and its connections with other metabolic pathways in the pathophysiology of Francisella novicida. The involvement of the PPP in the intracellular life cycle of Francisella was first demonstrated by studying PPP inactivating mutants. Indeed, we observed that inactivation of the tktA, rpiA or rpe genes severely impaired intramacrophage multiplication during the first 24 hours. However, time-lapse video microscopy demonstrated that rpiA and rpe mutants were able to resume late intracellular multiplication. To better understand the links between PPP and other metabolic networks in the bacterium, we also performed an extensive proteo-metabolomic analysis of these mutants. We show that the PPP constitutes a major bacterial metabolic hub with multiple connections to glycolysis, the tricarboxylic acid cycle and other pathways, such as fatty acid degradation and sulfur metabolism. Altogether our study highlights how PPP plays a key role in the pathogenesis and growth of Francisella in its intracellular niche.
Conflict of interest statement
No - The authors have declared that no competing interests exist.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

