Abnormality in the NK-cell population is prolonged in severe COVID-19 patients
- PMID: 34339730
- PMCID: PMC8324384
- DOI: 10.1016/j.jaci.2021.07.022
Abnormality in the NK-cell population is prolonged in severe COVID-19 patients
Abstract
Background: Our understanding of adaptive immune responses in patients with coronavirus disease 2019 (COVID-19) is rapidly evolving, but information on the innate immune responses by natural killer (NK) cells is still insufficient.
Objective: We aimed to examine the phenotypic and functional status of NK cells and their changes during the course of mild and severe COVID-19.
Methods: We performed RNA sequencing and flow cytometric analysis of NK cells from patients with mild and severe COVID-19 at multiple time points in the course of the disease using cryopreserved PBMCs.
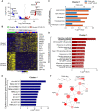
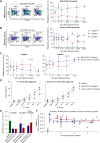
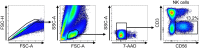
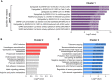
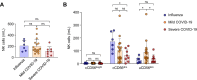
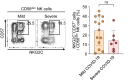
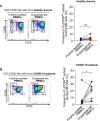
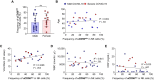
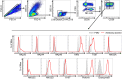
Results: In RNA-sequencing analysis, the NK cells exhibited distinctive features compared with healthy donors, with significant enrichment of proinflammatory cytokine-mediated signaling pathways. Intriguingly, we found that the unconventional CD56dimCD16neg NK-cell population expanded in cryopreserved PBMCs from patients with COVID-19 regardless of disease severity, accompanied by decreased NK-cell cytotoxicity. The NK-cell population was rapidly normalized alongside the disappearance of unconventional CD56dimCD16neg NK cells and the recovery of NK-cell cytotoxicity in patients with mild COVID-19, but this occurred slowly in patients with severe COVID-19.
Conclusions: The current longitudinal study provides a deep understanding of the NK-cell biology in COVID-19.
Keywords: COVID-19; NK cells; SARS-CoV-2; cytotoxicity; innate immunity; unconventional CD56(dim)CD16(neg) (uCD56(dim)) NK cell.
Copyright © 2021 American Academy of Allergy, Asthma & Immunology. Published by Elsevier Inc. All rights reserved.
Figures








References
-
- World Health Organization Weekly epidemiological update—19 January 2021. https://www.who.int/publications/m/item/weekly-epidemiological-update---... Available at:
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

