Decoding Post-Translational Modification Crosstalk With Proteomics
- PMID: 34339852
- PMCID: PMC8430371
- DOI: 10.1016/j.mcpro.2021.100129
Decoding Post-Translational Modification Crosstalk With Proteomics
Abstract
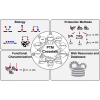
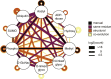
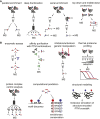
Post-translational modification (PTM) of proteins allows cells to regulate protein functions, transduce signals and respond to perturbations. PTMs expand protein functionality and diversity, which leads to increased proteome complexity. PTM crosstalk describes the combinatorial action of multiple PTMs on the same or on different proteins for higher order regulation. Here we review how recent advances in proteomic technologies, mass spectrometry instrumentation, and bioinformatics spurred the proteome-wide identification of PTM crosstalk through measurements of PTM sites. We provide an overview of the basic modes of PTM crosstalk, the proteomic methods to elucidate PTM crosstalk, and approaches that can inform about the functional consequences of PTM crosstalk.
Keywords: crosstalk; mass spectrometry; post-translational modification; proteomics.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


References
-
- Walsh C.T., Garneau-Tsodikova S., Gatto G.J. Protein posttranslational modifications: The chemistry of proteome diversifications. Angew. Chem. Int. Ed. Engl. 2005;44:7342–7372. - PubMed
-
- Sharma K., D’Souza R.C.J., Tyanova S., Schaab C., Wiśniewski J.R., Cox J., Mann M. Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thr-based signaling. Cell Rep. 2014;8:1583–1594. - PubMed
-
- Humphrey S.J., Azimifar S.B., Mann M. High-throughput phosphoproteomics reveals in vivo insulin signaling dynamics. Nat. Biotechnol. 2015;33:990–995. - PubMed
-
- Venne A.S., Kollipara L., Zahedi R.P. The next level of complexity: Crosstalk of posttranslational modifications. Proteomics. 2014;14:513–524. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

