Orientia tsutsugamushi Nucleomodulin Ank13 Exploits the RaDAR Nuclear Import Pathway To Modulate Host Cell Transcription
- PMID: 34340535
- PMCID: PMC8406279
- DOI: 10.1128/mBio.01816-21
Orientia tsutsugamushi Nucleomodulin Ank13 Exploits the RaDAR Nuclear Import Pathway To Modulate Host Cell Transcription
Abstract
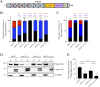
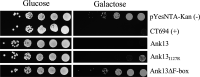
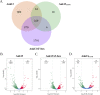
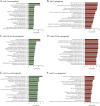
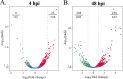
Orientia tsutsugamushi is the etiologic agent of scrub typhus, the deadliest of all diseases caused by obligate intracellular bacteria. Nucleomodulins, bacterial effectors that dysregulate eukaryotic transcription, are being increasingly recognized as key virulence factors. How they translocate into the nucleus and their functionally essential domains are poorly defined. We demonstrate that Ank13, an O. tsutsugamushi effector conserved among clinical isolates and expressed during infection, localizes to the nucleus in an importin β1-independent manner. Rather, Ank13 nucleotropism requires an isoleucine at the thirteenth position of its fourth ankyrin repeat, consistent with utilization of eukaryotic RaDAR (RanGDP-ankyrin repeats) nuclear import. RNA-seq analyses of cells expressing green fluorescent protein (GFP)-tagged Ank13, nucleotropism-deficient Ank13I127R, or Ank13ΔF-box, which lacks the F-box domain essential for interacting with SCF ubiquitin ligase, revealed Ank13 to be a nucleomodulin that predominantly downregulates transcription of more than 2,000 genes. Its ability to do so involves its nucleotropism and F-box in synergistic and mutually exclusive manners. Ank13 also acts in the cytoplasm to dysregulate smaller cohorts of genes. The effector's toxicity in yeast heavily depends on its F-box and less so on its nucleotropism. Genes negatively regulated by Ank13 include those involved in the inflammatory response, transcriptional control, and epigenetics. Importantly, the majority of genes that GFP-Ank13 most strongly downregulates are quiescent or repressed in O. tsutsugamushi-infected cells when Ank13 expression is strongest. Ank13 is the first nucleomodulin identified to coopt RaDAR and a multifaceted effector that functions in the nucleus and cytoplasm via F-box-dependent and -independent mechanisms to globally reprogram host cell transcription. IMPORTANCE Nucleomodulins are recently defined effectors used by diverse intracellular bacteria to manipulate eukaryotic gene expression and convert host cells into hospitable niches. How nucleomodulins enter the nucleus, their functional domains, and the genes that they modulate are incompletely characterized. Orientia tsutsugamushi is an intracellular bacterial pathogen that causes scrub typhus, which can be fatal. O. tsutsugamushi Ank13 is the first example of a microbial protein that coopts eukaryotic RaDAR (RanGDP-ankyrin repeats) nuclear import. It dysregulates expression of a multitude of host genes with those involved in transcriptional control and the inflammatory response being among the most prominent. Ank13 does so via mechanisms that are dependent and independent of both its nucleotropism and eukaryotic-like F-box domain that interfaces with ubiquitin ligase machinery. Nearly all the genes most strongly downregulated by ectopically expressed Ank13 are repressed in O. tsutsugamushi-infected cells, implicating its importance for intracellular colonization and scrub typhus molecular pathogenesis.
Keywords: Orientia tsutsugamushi; Rickettsia; ankyrin repeat; bacterial effector; intracellular bacterium; nucleomodulin.
Figures

Similar articles
-
Orientia tsutsugamushi Strain Ikeda Ankyrin Repeat-Containing Proteins Recruit SCF1 Ubiquitin Ligase Machinery via Poxvirus-Like F-Box Motifs.J Bacteriol. 2015 Oct;197(19):3097-109. doi: 10.1128/JB.00276-15. Epub 2015 Jul 13. J Bacteriol. 2015. PMID: 26170417 Free PMC article.
-
Multiple Orientia tsutsugamushi ankyrin repeat proteins interact with SCF1 ubiquitin ligase complex and eukaryotic elongation factor 1 α.PLoS One. 2014 Aug 28;9(8):e105652. doi: 10.1371/journal.pone.0105652. eCollection 2014. PLoS One. 2014. PMID: 25166298 Free PMC article.
-
Orientia tsutsugamushi uses two Ank effectors to modulate NF-κB p65 nuclear transport and inhibit NF-κB transcriptional activation.PLoS Pathog. 2018 May 7;14(5):e1007023. doi: 10.1371/journal.ppat.1007023. eCollection 2018 May. PLoS Pathog. 2018. PMID: 29734393 Free PMC article.
-
Orientia tsutsugamushi: An Unusual Intracellular Bacteria-Adaptation Strategies, Available Antibiotics, and Alternatives for Treatment.Curr Microbiol. 2024 Jun 21;81(8):236. doi: 10.1007/s00284-024-03754-1. Curr Microbiol. 2024. PMID: 38907107 Review.
-
Orientia tsutsugamushi DNA load and genotypes in blood as a marker of severity.Acta Trop. 2021 Mar;215:105786. doi: 10.1016/j.actatropica.2020.105786. Epub 2020 Dec 9. Acta Trop. 2021. PMID: 33309595 Review.
Cited by
-
Comparative virulence analysis of seven diverse strains of Orientia tsutsugamushi reveals a multifaceted and complex interplay of virulence factors responsible for disease.PLoS Pathog. 2025 Jun 30;21(6):e1012833. doi: 10.1371/journal.ppat.1012833. eCollection 2025 Jun. PLoS Pathog. 2025. PMID: 40587585 Free PMC article.
-
Orientia tsutsugamushi infection reduces host gluconeogenic but not glycolytic substrates.Infect Immun. 2024 Nov 12;92(11):e0028424. doi: 10.1128/iai.00284-24. Epub 2024 Sep 26. Infect Immun. 2024. PMID: 39324805 Free PMC article.
-
Orientia tsutsugamushi: analysis of the mobilome of a highly fragmented and repetitive genome reveals ongoing lateral gene transfer in an obligate intracellular bacterium.bioRxiv [Preprint]. 2023 May 11:2023.05.11.540415. doi: 10.1101/2023.05.11.540415. bioRxiv. 2023. Update in: mSphere. 2023 Dec 20;8(6):e0026823. doi: 10.1128/msphere.00268-23. PMID: 37215039 Free PMC article. Updated. Preprint.
-
Orientia tsutsugamushi Modulates RIPK3 Cellular Levels but Does Not Inhibit Necroptosis.Pathogens. 2025 May 14;14(5):478. doi: 10.3390/pathogens14050478. Pathogens. 2025. PMID: 40430799 Free PMC article.
-
The Obligate Intracellular Bacterial Pathogen Anaplasma phagocytophilum Exploits Host Cell Multivesicular Body Biogenesis for Proliferation and Dissemination.mBio. 2022 Dec 20;13(6):e0296122. doi: 10.1128/mbio.02961-22. Epub 2022 Nov 21. mBio. 2022. PMID: 36409075 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

