Captive Common Marmosets (Callithrix jacchus) Are Colonized throughout Their Lives by a Community of Bifidobacterium Species with Species-Specific Genomic Content That Can Support Adaptation to Distinct Metabolic Niches
- PMID: 34340536
- PMCID: PMC8406136
- DOI: 10.1128/mBio.01153-21
Captive Common Marmosets (Callithrix jacchus) Are Colonized throughout Their Lives by a Community of Bifidobacterium Species with Species-Specific Genomic Content That Can Support Adaptation to Distinct Metabolic Niches
Abstract
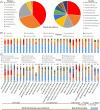
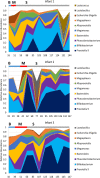
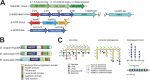
The common marmoset (Callithrix jacchus) is an omnivorous New World primate whose diet in the wild includes large amounts of fruit, seeds, flowers, and a variety of lizards and invertebrates. Marmosets also feed heavily on tree gums and exudates, and they have evolved unique morphological and anatomical characteristics to facilitate gum feeding (gummivory). In this study, we characterized the fecal microbiomes of adult and infant animals from a captive population of common marmosets at the Callitrichid Research Center at the University of Nebraska at Omaha under their normal dietary and environmental conditions. The microbiomes of adult animals were dominated by species of Bifidobacterium, Bacteroides, Prevotella, Phascolarctobacterium, Megamonas, and Megasphaera. Culturing and genomic analysis of the Bifidobacterium populations from adult animals identified four known marmoset-associated species (B. reuteri, B. aesculapii, B. myosotis, and B. hapali) and three unclassified taxa of Bifidobacterium that are phylogenetically distinct. Species-specific quantitative PCR (qPCR) confirmed that these same species of Bifidobacterium are abundant members of the microbiome throughout the lives of the animals. Genomic loci in each Bifidobacterium species encode enzymes to support growth and major marmoset milk oligosaccharides during breastfeeding; however, metabolic islands that can support growth on complex polysaccharide substrates in the diets of captive adults (pectin, xyloglucan, and xylan), including loci in B. aesculapii that can support its unique ability to grow on arabinogalactan-rich tree gums, were species-specific. IMPORTANCEBifidobacterium species are recognized as important, beneficial microbes in the human gut microbiome, and their ability colonize individuals at different stages of life is influenced by host, dietary, environmental, and ecological factors, which is poorly understood. The common marmoset is an emerging nonhuman primate model with a short maturation period, making this model amenable to study the microbiome throughout a life history. Features of the microbiome in captive marmosets are also shared with human gut microbiomes, including abundant populations of Bifidobacterium species. Our studies show that several species of Bifidobacterium are dominant members of the captive marmoset microbiome throughout their life history. Metabolic capacities in genomes of the marmoset Bifidobacterium species suggest species-specific adaptations to different components of the captive marmoset diet, including the unique capacity in B. aesculapii for degradation of gum arabic, suggesting that regular dietary exposure in captivity may be important for preserving gum-degrading species in the microbiome.
Keywords: Bifidobacterium; common marmoset; diet-driven gut microbiomes; gum arabic; gum-feeding specialists.
Figures






Similar articles
-
The gut microbiome of exudivorous marmosets in the wild and captivity.Sci Rep. 2022 Mar 23;12(1):5049. doi: 10.1038/s41598-022-08797-7. Sci Rep. 2022. PMID: 35322053 Free PMC article.
-
Impact of diet change on the gut microbiome of common marmosets (Callithrix jacchus).mSystems. 2024 Aug 20;9(8):e0010824. doi: 10.1128/msystems.00108-24. Epub 2024 Jul 8. mSystems. 2024. PMID: 38975760 Free PMC article.
-
The Gastrointestinal Microbiota of the Common Marmoset (Callithrix jacchus).ILAR J. 2020 Dec 31;61(2-3):188-198. doi: 10.1093/ilar/ilaa025. ILAR J. 2020. PMID: 33620078 Free PMC article. Review.
-
Comparative genomics of Bifidobacterium species isolated from marmosets and humans.Am J Primatol. 2019 Oct;81(10-11):e983. doi: 10.1002/ajp.22983. Epub 2019 May 6. Am J Primatol. 2019. PMID: 31062394 Free PMC article.
-
Varied Pathways of Infant Gut-Associated Bifidobacterium to Assimilate Human Milk Oligosaccharides: Prevalence of the Gene Set and Its Correlation with Bifidobacteria-Rich Microbiota Formation.Nutrients. 2019 Dec 26;12(1):71. doi: 10.3390/nu12010071. Nutrients. 2019. PMID: 31888048 Free PMC article. Review.
Cited by
-
The gut microbiome of exudivorous marmosets in the wild and captivity.Sci Rep. 2022 Mar 23;12(1):5049. doi: 10.1038/s41598-022-08797-7. Sci Rep. 2022. PMID: 35322053 Free PMC article.
-
Antibiotic-induced gut dysbiosis elicits gut-brain axis relevant multi-omic signatures and behavioral and neuroendocrine changes in a nonhuman primate model.Gut Microbes. 2024 Jan-Dec;16(1):2305476. doi: 10.1080/19490976.2024.2305476. Epub 2024 Jan 29. Gut Microbes. 2024. PMID: 38284649 Free PMC article.
-
Impact of diet change on the gut microbiome of common marmosets (Callithrix jacchus).mSystems. 2024 Aug 20;9(8):e0010824. doi: 10.1128/msystems.00108-24. Epub 2024 Jul 8. mSystems. 2024. PMID: 38975760 Free PMC article.
-
Integrative genomic reconstruction reveals heterogeneity in carbohydrate utilization across human gut bifidobacteria.Nat Microbiol. 2025 Aug;10(8):2031-2047. doi: 10.1038/s41564-025-02056-x. Epub 2025 Jul 16. Nat Microbiol. 2025. PMID: 40670725 Free PMC article.
-
Integrative genomic reconstruction reveals heterogeneity in carbohydrate utilization across human gut bifidobacteria.bioRxiv [Preprint]. 2025 May 17:2024.07.06.602360. doi: 10.1101/2024.07.06.602360. bioRxiv. 2025. Update in: Nat Microbiol. 2025 Aug;10(8):2031-2047. doi: 10.1038/s41564-025-02056-x. PMID: 39005317 Free PMC article. Updated. Preprint.
References
-
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto J-M, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Li S, Jian M, Zhou Y, Li Y, Zhang X, Li S, Qin N, Yang H, Wang J, Brunak S, Doré J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, MetaHIT Consortium , et al.. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59–65. doi:10.1038/nature08821. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
