In vitro turnover numbers do not reflect in vivo activities of yeast enzymes
- PMID: 34341111
- PMCID: PMC8364156
- DOI: 10.1073/pnas.2108391118
In vitro turnover numbers do not reflect in vivo activities of yeast enzymes
Abstract
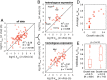
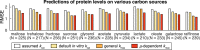
Turnover numbers (kcat values) quantitatively represent the activity of enzymes, which are mostly measured in vitro. While a few studies have reported in vivo catalytic rates (kapp values) in bacteria, a large-scale estimation of kapp in eukaryotes is lacking. Here, we estimated kapp of the yeast Saccharomyces cerevisiae under diverse conditions. By comparing the maximum kapp across conditions with in vitro kcat we found a weak correlation in log scale of R2 = 0.28, which is lower than for Escherichia coli (R2 = 0.62). The weak correlation is caused by the fact that many in vitro kcat values were measured for enzymes obtained through heterologous expression. Removal of these enzymes improved the correlation to R2 = 0.41 but still not as good as for E. coli, suggesting considerable deviations between in vitro and in vivo enzyme activities in yeast. By parameterizing an enzyme-constrained metabolic model with our kapp dataset we observed better performance than the default model with in vitro kcat in predicting proteomics data, demonstrating the strength of using the dataset generated here.
Keywords: Saccharomyces cerevisiae; kcat; metabolism; proteomics; turnover number.
Copyright © 2021 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures
Similar articles
-
Yeast9: a consensus genome-scale metabolic model for S. cerevisiae curated by the community.Mol Syst Biol. 2024 Oct;20(10):1134-1150. doi: 10.1038/s44320-024-00060-7. Epub 2024 Aug 12. Mol Syst Biol. 2024. PMID: 39134886 Free PMC article.
-
Comparison of the small molecule metabolic enzymes of Escherichia coli and Saccharomyces cerevisiae.Genome Res. 2002 Jun;12(6):916-29. doi: 10.1101/gr.228002. Genome Res. 2002. PMID: 12045145 Free PMC article.
-
Genome-scale modeling of the protein secretory machinery in yeast.PLoS One. 2013 May 7;8(5):e63284. doi: 10.1371/journal.pone.0063284. Print 2013. PLoS One. 2013. PMID: 23667601 Free PMC article.
-
The importance of post-translational modifications in regulating Saccharomyces cerevisiae metabolism.FEMS Yeast Res. 2012 Mar;12(2):104-17. doi: 10.1111/j.1567-1364.2011.00765.x. Epub 2011 Dec 22. FEMS Yeast Res. 2012. PMID: 22128902 Review.
-
Glucose repression in Saccharomyces cerevisiae.FEMS Yeast Res. 2015 Sep;15(6):fov068. doi: 10.1093/femsyr/fov068. Epub 2015 Jul 22. FEMS Yeast Res. 2015. PMID: 26205245 Free PMC article. Review.
Cited by
-
Data integration across conditions improves turnover number estimates and metabolic predictions.Nat Commun. 2023 Mar 17;14(1):1485. doi: 10.1038/s41467-023-37151-2. Nat Commun. 2023. PMID: 36932067 Free PMC article.
-
Genome-scale modeling of yeast metabolism: retrospectives and perspectives.FEMS Yeast Res. 2022 Feb 22;22(1):foac003. doi: 10.1093/femsyr/foac003. FEMS Yeast Res. 2022. PMID: 35094064 Free PMC article. Review.
-
Dynamic Cytoplasm: A Physical Regulator of Enzymatic Function.Biochemistry. 2025 Jul 1;64(13):2699-2711. doi: 10.1021/acs.biochem.5c00212. Epub 2025 Jun 15. Biochemistry. 2025. PMID: 40517322 Free PMC article. Review.
-
High-throughput and reliable acquisition of in vivo turnover number fuels precise metabolic engineering.Synth Syst Biotechnol. 2022 Jan 5;7(1):541-543. doi: 10.1016/j.synbio.2021.12.006. eCollection 2022 Mar. Synth Syst Biotechnol. 2022. PMID: 35059513 Free PMC article.
-
PARROT: Prediction of enzyme abundances using protein-constrained metabolic models.PLoS Comput Biol. 2023 Oct 19;19(10):e1011549. doi: 10.1371/journal.pcbi.1011549. eCollection 2023 Oct. PLoS Comput Biol. 2023. PMID: 37856550 Free PMC article.
References
-
- Davidi D., Milo R., Lessons on enzyme kinetics from quantitative proteomics. Curr. Opin. Biotechnol. 46, 81–89 (2017). - PubMed
-
- Lahtvee P.-J., et al. ., Absolute quantification of protein and mRNA abundances demonstrate variability in gene-specific translation efficiency in yeast. Cell Syst. 4, 495–504.e5 (2017). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

