A machine learning case-control classifier for schizophrenia based on DNA methylation in blood
- PMID: 34341337
- PMCID: PMC8329061
- DOI: 10.1038/s41398-021-01496-3
A machine learning case-control classifier for schizophrenia based on DNA methylation in blood
Abstract
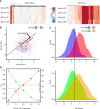
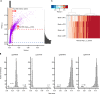
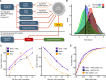
Epigenetic dysregulation is thought to contribute to the etiology of schizophrenia (SZ), but the cell type-specificity of DNA methylation makes population-based epigenetic studies of SZ challenging. To train an SZ case-control classifier based on DNA methylation in blood, therefore, we focused on human genomic regions of systemic interindividual epigenetic variation (CoRSIVs), a subset of which are represented on the Illumina Human Methylation 450K (HM450) array. HM450 DNA methylation data on whole blood of 414 SZ cases and 433 non-psychiatric controls were used as training data for a classification algorithm with built-in feature selection, sparse partial least squares discriminate analysis (SPLS-DA); application of SPLS-DA to HM450 data has not been previously reported. Using the first two SPLS-DA dimensions we calculated a "risk distance" to identify individuals with the highest probability of SZ. The model was then evaluated on an independent HM450 data set on 353 SZ cases and 322 non-psychiatric controls. Our CoRSIV-based model classified 303 individuals as cases with a positive predictive value (PPV) of 80%, far surpassing the performance of a model based on polygenic risk score (PRS). Importantly, risk distance (based on CoRSIV methylation) was not associated with medication use, arguing against reverse causality. Risk distance and PRS were positively correlated (Pearson r = 0.28, P = 1.28 × 10-12), and mediational analysis suggested that genetic effects on SZ are partially mediated by altered methylation at CoRSIVs. Our results indicate two innate dimensions of SZ risk: one based on genetic, and the other on systemic epigenetic variants.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

