Nuclear compartmentalization as a mechanism of quantitative control of gene expression
- PMID: 34341548
- PMCID: PMC12145136
- DOI: 10.1038/s41580-021-00387-1
Nuclear compartmentalization as a mechanism of quantitative control of gene expression
Abstract
Gene regulation requires the dynamic coordination of hundreds of regulatory factors at precise genomic and RNA targets. Although many regulatory factors have specific affinity for their nucleic acid targets, molecular diffusion and affinity models alone cannot explain many of the quantitative features of gene regulation in the nucleus. One emerging explanation for these quantitative properties is that DNA, RNA and proteins organize within precise, 3D compartments in the nucleus to concentrate groups of functionally related molecules. Recently, nucleic acids and proteins involved in many important nuclear processes have been shown to engage in cooperative interactions, which lead to the formation of condensates that partition the nucleus. In this Review, we discuss an emerging perspective of gene regulation, which moves away from classic models of stoichiometric interactions towards an understanding of how spatial compartmentalization can lead to non-stoichiometric molecular interactions and non-linear regulatory behaviours. We describe key mechanisms of nuclear compartment formation, including emerging roles for non-coding RNAs in facilitating their formation, and discuss the functional role of nuclear compartments in transcription regulation, co-transcriptional and post-transcriptional RNA processing, and higher-order chromatin regulation. More generally, we discuss how compartmentalization may explain important quantitative aspects of gene regulation.
© 2021. Springer Nature Limited.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures

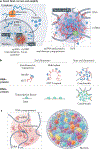
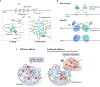



References
-
- Mirny L et al. How a protein searches for its site on DNA: the mechanism of facilitated diffusion. J. Phys. A Math. Theor. 42, 434013 (2009).
-
- Jana T, Brodsky S & Barkai N Speed–specificity trade-offs in the transcription factors search for their genomic binding sites. Trends Genet. 37, 421–432 (2021). - PubMed
-
- Pombo A & Dillon N Three-dimensional genome architecture: players and mechanisms. Nat. Rev. Mol. Cell Biol. 16, 245–257 (2015). - PubMed
-
- Bonev B & Cavalli G Organization and function of the 3D genome. Nat. Rev. Genet. 17, 772–772 (2016). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

