This is a preprint.
Gut microbiome dysbiosis during COVID-19 is associated with increased risk for bacteremia and microbial translocation
- PMID: 34341786
- PMCID: PMC8328072
- DOI: 10.21203/rs.3.rs-726620/v1
Gut microbiome dysbiosis during COVID-19 is associated with increased risk for bacteremia and microbial translocation
Update in
-
Gut microbiome dysbiosis in antibiotic-treated COVID-19 patients is associated with microbial translocation and bacteremia.Nat Commun. 2022 Nov 1;13(1):5926. doi: 10.1038/s41467-022-33395-6. Nat Commun. 2022. PMID: 36319618 Free PMC article.
Abstract
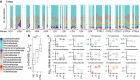
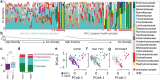
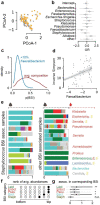
The microbial populations in the gut microbiome have recently been associated with COVID-19 disease severity. However, a causal impact of the gut microbiome on COVID-19 patient health has not been established. Here we provide evidence that gut microbiome dysbiosis is associated with translocation of bacteria into the blood during COVID-19, causing life-threatening secondary infections. Antibiotics and other treatments during COVID-19 can potentially confound microbiome associations. We therefore first demonstrate that the gut microbiome is directly affected by SARS-CoV-2 infection in a dose-dependent manner in a mouse model, causally linking viral infection and gut microbiome dysbiosis. Comparison with stool samples collected from 97 COVID-19 patients at two different clinical sites also revealed substantial gut microbiome dysbiosis, paralleling our observations in the animal model. Specifically, we observed blooms of opportunistic pathogenic bacterial genera known to include antimicrobial-resistant species in hospitalized COVID-19 patients. Analysis of blood culture results testing for secondary microbial bloodstream infections with paired microbiome data obtained from these patients suggest that bacteria translocate from the gut into the systemic circulation of COVID-19 patients. These results are consistent with a direct role for gut microbiome dysbiosis in enabling dangerous secondary infections during COVID 19.
Figures
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

