Catching SARS-CoV-2 by Sequence Hybridization: a Comparative Analysis
- PMID: 34342536
- PMCID: PMC8407296
- DOI: 10.1128/mSystems.00392-21
Catching SARS-CoV-2 by Sequence Hybridization: a Comparative Analysis
Abstract
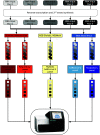
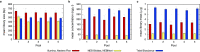
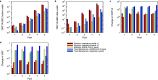
Controlling and monitoring the still ongoing severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic regarding geographical distribution, evolution, and emergence of new mutations of the SARS-CoV-2 virus is only possible due to continuous next-generation sequencing (NGS) and sharing sequence data worldwide. Efficient sequencing strategies enable the retrieval of increasing numbers of high-quality, full-length genomes and are, hence, indispensable. Two opposed enrichment methods, tiling multiplex PCR and sequence hybridization by bait capture, have been established for SARS-CoV-2 sequencing and are both frequently used, depending on the quality of the patient sample and the question at hand. Here, we focused on the evaluation of the sequence hybridization method by studying five commercially available sequence capture bait panels with regard to sensitivity and capture efficiency. We discovered the SARS-CoV-2-specific panel of Twist Bioscience to be the most efficient panel, followed by two respiratory panels from Twist Bioscience and Illumina, respectively. Our results provide on the one hand a decision basis for the sequencing community including a computation for using the full capacity of the flow cell and on the other hand potential improvements for the manufacturers. IMPORTANCE Sequencing the genomes of the circulating SARS-CoV-2 strains is the only way to monitor the viral spread and evolution of the virus. Two different approaches, namely, tiling multiplex PCR and sequence hybridization by bait capture, are commonly used to fulfill this task. This study describes for the first time a combined approach of droplet digital PCR (ddPCR) and NGS to evaluate five commercially available sequence capture panels targeting SARS-CoV-2. In doing so, we were able to determine the most sensitive and efficient capture panel, distinguish the mode of action of the various bait panels, and compute the number of read pairs needed to recover a high-quality full-length genome. By calculating the minimum number of read pairs needed, we are providing optimized flow cell loading conditions for all sequencing laboratories worldwide that are striving for maximizing sequencing output and simultaneously minimizing time, costs, and sequencing resources.
Keywords: NGS; SARS-CoV-2; adaptive mutations; ddPCR; enrichment; mutations; next-generation sequencing; sequence capture.
Figures

References
-
- Wu F, Zhao S, Yu B, Chen Y-M, Wang W, Song Z-G, Hu Y, Tao Z-W, Tian J-H, Pei Y-Y, Yuan M-L, Zhang Y-L, Dai F-H, Liu Y, Wang Q-M, Zheng J-J, Xu L, Holmes EC, Zhang Y-Z. 2020. A new coronavirus associated with human respiratory disease in China. Nature 579:265–269. doi:10.1038/s41586-020-2008-3. - DOI - PMC - PubMed
-
- Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team . 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. doi:10.1056/NEJMoa2001017. - DOI - PMC - PubMed
-
- Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y, Ma X, Zhan F, Wang L, Hu T, Zhou H, Hu Z, Zhou W, Zhao L, Chen J, Meng Y, Wang J, Lin Y, Yuan J, Xie Z, Ma J, Liu WJ, Wang D, Xu W, Holmes EC, Gao GF, Wu G, Chen W, Shi W, Tan W. 2020. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395:565–574. doi:10.1016/S0140-6736(20)30251-8. - DOI - PMC - PubMed
-
- Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, Bleicker T, Brünink S, Schneider J, Schmidt ML, Mulders DG, Haagmans BL, van der Veer B, van den Brink S, Wijsman L, Goderski G, Romette J-L, Ellis J, Zambon M, Peiris M, Goossens H, Reusken C, Koopmans MP, Drosten C. 2020. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 25:2000045. doi:10.2807/1560-7917.ES.2020.25.3.2000045. - DOI - PMC - PubMed
-
- Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, Hengartner N, Giorgi EE, Bhattacharya T, Foley B, Hastie KM, Parker MD, Partridge DG, Evans CM, Freeman TM, de Silva TI, McDanal C, Perez LG, Tang H, Moon-Walker A, Whelan SP, LaBranche CC, Saphire EO, Montefiori DC, Sheffield COVID-19 Genomics Group . 2020. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell 182:812–827.e19. doi:10.1016/j.cell.2020.06.043. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

