Mitochondrial RNase H1 activity regulates R-loop homeostasis to maintain genome integrity and enable early embryogenesis in Arabidopsis
- PMID: 34343166
- PMCID: PMC8330923
- DOI: 10.1371/journal.pbio.3001357
Mitochondrial RNase H1 activity regulates R-loop homeostasis to maintain genome integrity and enable early embryogenesis in Arabidopsis
Abstract
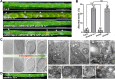
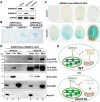
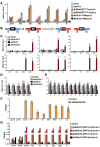
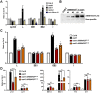
Plant mitochondrial genomes undergo frequent homologous recombination (HR). Ectopic HR activity is inhibited by the HR surveillance pathway, but the underlying regulatory mechanism is unclear. Here, we show that the mitochondrial RNase H1 AtRNH1B impairs the formation of RNA:DNA hybrids (R-loops) and participates in the HR surveillance pathway in Arabidopsis thaliana. AtRNH1B suppresses ectopic HR at intermediate-sized repeats (IRs) and thus maintains mitochondrial DNA (mtDNA) replication. The RNase H1 AtRNH1C is restricted to the chloroplast; however, when cells lack AtRNH1B, transport of chloroplast AtRNH1C into the mitochondria secures HR surveillance, thus ensuring the integrity of the mitochondrial genome and allowing embryogenesis to proceed. HR surveillance is further regulated by the single-stranded DNA-binding protein ORGANELLAR SINGLE-STRANDED DNA BINDING PROTEIN1 (OSB1), which decreases the formation of R-loops. This study uncovers a facultative dual targeting mechanism between organelles and sheds light on the roles of RNase H1 in organellar genome maintenance and embryogenesis.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





Similar articles
-
RNase H1 Cooperates with DNA Gyrases to Restrict R-Loops and Maintain Genome Integrity in Arabidopsis Chloroplasts.Plant Cell. 2017 Oct;29(10):2478-2497. doi: 10.1105/tpc.17.00305. Epub 2017 Sep 22. Plant Cell. 2017. PMID: 28939594 Free PMC article.
-
RNase H1C collaborates with ssDNA binding proteins WHY1/3 and recombinase RecA1 to fulfill the DNA damage repair in Arabidopsis chloroplasts.Nucleic Acids Res. 2021 Jul 9;49(12):6771-6787. doi: 10.1093/nar/gkab479. Nucleic Acids Res. 2021. PMID: 34133716 Free PMC article.
-
Evolutionary History and Activity of RNase H1-Like Proteins in Arabidopsis thaliana.Plant Cell Physiol. 2020 Jun 1;61(6):1107-1119. doi: 10.1093/pcp/pcaa040. Plant Cell Physiol. 2020. PMID: 32191307 Free PMC article.
-
The Jekyll and Hyde character of RNase H1 and its multiple roles in mitochondrial DNA metabolism.DNA Repair (Amst). 2019 Dec;84:102630. doi: 10.1016/j.dnarep.2019.06.001. Epub 2019 Jun 4. DNA Repair (Amst). 2019. PMID: 31178343 Review.
-
RNase H1, the Gold Standard for R-Loop Detection.Methods Mol Biol. 2022;2528:91-114. doi: 10.1007/978-1-0716-2477-7_7. Methods Mol Biol. 2022. PMID: 35704187 Review.
Cited by
-
Walking a tightrope: The complex balancing act of R-loops in genome stability.Mol Cell. 2022 Jun 16;82(12):2267-2297. doi: 10.1016/j.molcel.2022.04.014. Epub 2022 May 3. Mol Cell. 2022. PMID: 35508167 Free PMC article. Review.
-
How RNases Shape Mitochondrial Transcriptomes.Int J Mol Sci. 2022 May 30;23(11):6141. doi: 10.3390/ijms23116141. Int J Mol Sci. 2022. PMID: 35682820 Free PMC article. Review.
-
Ribonucleotide and R-Loop Damage in Plastid DNA and Mitochondrial DNA during Maize Development.Plants (Basel). 2023 Sep 2;12(17):3161. doi: 10.3390/plants12173161. Plants (Basel). 2023. PMID: 37687407 Free PMC article.
-
Regulation of Heat Stress in Physcomitrium (Physcomitrella) patens Provides Novel Insight into the Functions of Plant RNase H1s.Int J Mol Sci. 2022 Aug 17;23(16):9270. doi: 10.3390/ijms23169270. Int J Mol Sci. 2022. PMID: 36012542 Free PMC article.
-
Comparative organelle genomics in Daphniphyllaceae reveal phylogenetic position and organelle structure evolution.BMC Genomics. 2025 Jan 15;26(1):40. doi: 10.1186/s12864-025-11213-9. BMC Genomics. 2025. PMID: 39815181 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

