Distributed hybrid-indexing of compressed pan-genomes for scalable and fast sequence alignment
- PMID: 34343181
- PMCID: PMC8330939
- DOI: 10.1371/journal.pone.0255260
Distributed hybrid-indexing of compressed pan-genomes for scalable and fast sequence alignment
Abstract
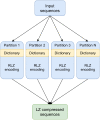
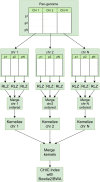
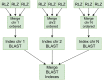
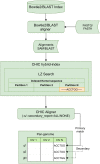
Computational pan-genomics utilizes information from multiple individual genomes in large-scale comparative analysis. Genetic variation between case-controls, ethnic groups, or species can be discovered thoroughly using pan-genomes of such subpopulations. Whole-genome sequencing (WGS) data volumes are growing rapidly, making genomic data compression and indexing methods very important. Despite current space-efficient repetitive sequence compression and indexing methods, the deployed compression methods are often sequential, computationally time-consuming, and do not provide efficient sequence alignment performance on vast collections of genomes such as pan-genomes. For performing rapid analytics with the ever-growing genomics data, data compression and indexing methods have to exploit distributed and parallel computing more efficiently. Instead of strict genome data compression methods, we will focus on the efficient construction of a compressed index for pan-genomes. Compressed hybrid-index enables fast sequence alignments to several genomes at once while shrinking the index size significantly compared to traditional indexes. We propose a scalable distributed compressed hybrid-indexing method for large genomic data sets enabling pan-genome-based sequence search and read alignment capabilities. We show the scalability of our tool, DHPGIndex, by executing experiments in a distributed Apache Spark-based computing cluster comprising 448 cores distributed over 26 nodes. The experiments have been performed both with human and bacterial genomes. DHPGIndex built a BLAST index for n = 250 human pan-genome with an 870:1 compression ratio (CR) in 342 minutes and a Bowtie2 index with 157:1 CR in 397 minutes. For n = 1,000 human pan-genome, the BLAST index was built in 1520 minutes with 532:1 CR and the Bowtie2 index in 1938 minutes with 76:1 CR. Bowtie2 aligned 14.6 GB of paired-end reads to the compressed (n = 1,000) index in 31.7 minutes on a single node. Compressing n = 13,375,031 (488 GB) GenBank database to BLAST index resulted in CR of 62:1 in 575 minutes. BLASTing 189,864 Crispr-Cas9 gRNA target sequences (23 MB in total) to the compressed index of human pan-genome (n = 1,000) finished in 45 minutes on a single node. 30 MB mixed bacterial sequences were (n = 599) were blasted to the compressed index of 488 GB GenBank database (n = 13,375,031) in 26 minutes on 25 nodes. 78 MB mixed sequences (n = 4,167) were blasted to the compressed index of 18 GB E. coli sequence database (n = 745,409) in 5.4 minutes on a single node.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Efficient Construction of a Complete Index for Pan-Genomics Read Alignment.J Comput Biol. 2020 Apr;27(4):500-513. doi: 10.1089/cmb.2019.0309. Epub 2020 Mar 16. J Comput Biol. 2020. PMID: 32181684 Free PMC article.
-
ViraPipe: scalable parallel pipeline for viral metagenome analysis from next generation sequencing reads.Bioinformatics. 2018 Mar 15;34(6):928-935. doi: 10.1093/bioinformatics/btx702. Bioinformatics. 2018. PMID: 29106455
-
Large-scale compression of genomic sequence databases with the Burrows-Wheeler transform.Bioinformatics. 2012 Jun 1;28(11):1415-9. doi: 10.1093/bioinformatics/bts173. Epub 2012 May 3. Bioinformatics. 2012. PMID: 22556365
-
Insights from 20 years of bacterial genome sequencing.Funct Integr Genomics. 2015 Mar;15(2):141-61. doi: 10.1007/s10142-015-0433-4. Epub 2015 Feb 27. Funct Integr Genomics. 2015. PMID: 25722247 Free PMC article. Review.
-
Optimizing the Parametrization of Homologue Classification in the Pan-Genome Computation for a Bacterial Species: Case Study Streptococcus pyogenes.Methods Mol Biol. 2022;2449:299-324. doi: 10.1007/978-1-0716-2095-3_13. Methods Mol Biol. 2022. PMID: 35507269 Review.
Cited by
-
Framing Apache Spark in life sciences.Heliyon. 2023 Feb 9;9(2):e13368. doi: 10.1016/j.heliyon.2023.e13368. eCollection 2023 Feb. Heliyon. 2023. PMID: 36852030 Free PMC article. Review.
References
-
- National Human Genome Research Institute. The Cost of Sequencing a Human Genome. 2020. Available from: https://www.genome.gov/sequencingcosts/
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

