Dynamic cell contacts between periportal mesenchyme and ductal epithelium act as a rheostat for liver cell proliferation
- PMID: 34343491
- PMCID: PMC8577825
- DOI: 10.1016/j.stem.2021.07.002
Dynamic cell contacts between periportal mesenchyme and ductal epithelium act as a rheostat for liver cell proliferation
Abstract
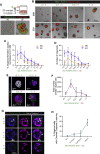
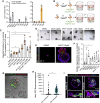
In the liver, ductal cells rarely proliferate during homeostasis but do so transiently after tissue injury. These cells can be expanded as organoids that recapitulate several of the cell-autonomous mechanisms of regeneration but lack the stromal interactions of the native tissue. Here, using organoid co-cultures that recapitulate the ductal-to-mesenchymal cell architecture of the portal tract, we demonstrate that a subpopulation of mouse periportal mesenchymal cells exerts dual control on proliferation of the epithelium. Ductal cell proliferation is either induced and sustained or, conversely, completely abolished, depending on the number of direct mesenchymal cell contacts, through a mechanism mediated, at least in part, by Notch signaling. Our findings expand the concept of the cellular niche in epithelial tissues, whereby not only soluble factors but also cell-cell contacts are the key regulatory cues involved in the control of cellular behaviors, suggesting a critical role for cell-cell contacts during regeneration.
Keywords: droplet microfluidics; flow-focussing device; liver; liver ductal cell; mesenchyme; multicellular co-culture; niche; organoid; organotypic co-culture; regeneration.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests M.H. is inventor in a patent on liver organoids and is on the advisory board of the journal Cell Stem Cell.
Figures






Comment in
-
Can't touch this: Stromal-mediated ductal proliferation.Cell Stem Cell. 2021 Nov 4;28(11):1885-1887. doi: 10.1016/j.stem.2021.10.006. Cell Stem Cell. 2021. PMID: 34739828
References
-
- Aloia L., McKie M.A., Vernaz G., Cordero-Espinoza L., Aleksieva N., van den Ameele J., Antonica F., Font-Cunill B., Raven A., Aiese Cigliano R., et al. Epigenetic remodelling licences adult cholangiocytes for organoid formation and liver regeneration. Nat. Cell Biol. 2019;21:1321–1333. - PMC - PubMed
-
- Apte U., Thompson M.D., Cui S., Liu B., Cieply B., Monga S.P. Wnt/beta-catenin signaling mediates oval cell response in rodents. Hepatology. 2008;47:288–295. - PubMed
-
- Berger D.R., Ware B.R., Davidson M.D., Allsup S.R., Khetani S.R. Enhancing the functional maturity of induced pluripotent stem cell-derived human hepatocytes by controlled presentation of cell-cell interactions in vitro. Hepatology. 2015;61:1370–1381. - PubMed
-
- Bhatia S.N., Balis U.J., Yarmush M.L., Toner M. Microfabrication of hepatocyte/fibroblast co-cultures: role of homotypic cell interactions. Biotechnol. Prog. 1998;14:378–387. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

