Pattern recognition receptors in health and diseases
- PMID: 34344870
- PMCID: PMC8333067
- DOI: 10.1038/s41392-021-00687-0
Pattern recognition receptors in health and diseases
Abstract
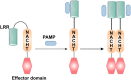
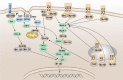
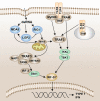
Pattern recognition receptors (PRRs) are a class of receptors that can directly recognize the specific molecular structures on the surface of pathogens, apoptotic host cells, and damaged senescent cells. PRRs bridge nonspecific immunity and specific immunity. Through the recognition and binding of ligands, PRRs can produce nonspecific anti-infection, antitumor, and other immunoprotective effects. Most PRRs in the innate immune system of vertebrates can be classified into the following five types based on protein domain homology: Toll-like receptors (TLRs), nucleotide oligomerization domain (NOD)-like receptors (NLRs), retinoic acid-inducible gene-I (RIG-I)-like receptors (RLRs), C-type lectin receptors (CLRs), and absent in melanoma-2 (AIM2)-like receptors (ALRs). PRRs are basically composed of ligand recognition domains, intermediate domains, and effector domains. PRRs recognize and bind their respective ligands and recruit adaptor molecules with the same structure through their effector domains, initiating downstream signaling pathways to exert effects. In recent years, the increased researches on the recognition and binding of PRRs and their ligands have greatly promoted the understanding of different PRRs signaling pathways and provided ideas for the treatment of immune-related diseases and even tumors. This review describes in detail the history, the structural characteristics, ligand recognition mechanism, the signaling pathway, the related disease, new drugs in clinical trials and clinical therapy of different types of PRRs, and discusses the significance of the research on pattern recognition mechanism for the treatment of PRR-related diseases.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

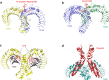

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

