Transcriptomic landscape of blood platelets in healthy donors
- PMID: 34344933
- PMCID: PMC8333095
- DOI: 10.1038/s41598-021-94003-z
Transcriptomic landscape of blood platelets in healthy donors
Abstract
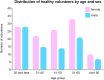
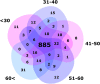
Blood platelet RNA-sequencing is increasingly used among the scientific community. Aberrant platelet transcriptome is common in cancer or cardiovascular disease, but reference data on platelet RNA content in healthy individuals are scarce and merit complex investigation. We sought to explore the dynamics of platelet transcriptome. Datasets from 204 healthy donors were used for the analysis of splice variants, particularly with regard to age, sex, blood storage time, unit of collection or library size. Genes B2M, PPBP, TMSB4X, ACTB, FTL, CLU, PF4, F13A1, GNAS, SPARC, PTMA, TAGLN2, OAZ1 and OST4 demonstrated the highest expression in the analysed cohort, remaining substantial transcription consistency. CSF3R gene was found upregulated in males (fold change 2.10, FDR q < 0.05). Cohort dichotomisation according to the median age, showed upregulated KSR1 in the older donors (fold change 2.11, FDR q < 0.05). Unsupervised hierarchical clustering revealed two clusters which were irrespective of age, sex, storage time, collecting unit or library size. However, when donors are analysed globally (as vectors), sex, storage time, library size, the unit of blood collection as well as age impose a certain degree of between- and/or within-group variability. Healthy donor platelet transcriptome retains general consistency, with very few splice variants deviating from the landscape. Although multidimensional analysis reveals statistically significant variability between and within the analysed groups, biologically, these changes are minor and irrelevant while considering disease classification. Our work provides a reference for studies working both on healthy platelets and pathological conditions affecting platelet transcriptome.
© 2021. The Author(s).
Conflict of interest statement
TW, MGB, MTR are inventors on relevant patents. TW is shareholder of GRAIL Inc. AS, MP, KP, PG, SIV, BS, NBK, TS, JJ, AJS have no competing interests.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

