A low Smc flux avoids collisions and facilitates chromosome organization in Bacillus subtilis
- PMID: 34346312
- PMCID: PMC8357415
- DOI: 10.7554/eLife.65467
A low Smc flux avoids collisions and facilitates chromosome organization in Bacillus subtilis
Abstract
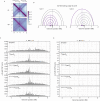
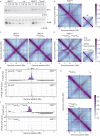
SMC complexes are widely conserved ATP-powered DNA-loop-extrusion motors indispensable for organizing and faithfully segregating chromosomes. How SMC complexes translocate along DNA for loop extrusion and what happens when two complexes meet on the same DNA molecule is largely unknown. Revealing the origins and the consequences of SMC encounters is crucial for understanding the folding process not only of bacterial, but also of eukaryotic chromosomes. Here, we uncover several factors that influence bacterial chromosome organization by modulating the probability of such clashes. These factors include the number, the strength, and the distribution of Smc loading sites, the residency time on the chromosome, the translocation rate, and the cellular abundance of Smc complexes. By studying various mutants, we show that these parameters are fine-tuned to reduce the frequency of encounters between Smc complexes, presumably as a risk mitigation strategy. Mild perturbations hamper chromosome organization by causing Smc collisions, implying that the cellular capacity to resolve them is limited. Altogether, we identify mechanisms that help to avoid Smc collisions and their resolution by Smc traversal or other potentially risky molecular transactions.
Keywords: B. subtilis; DNA loop extrusion; ParABS; SMC complexes; chromosome condensation; chromosome organization; chromosome segregation; chromosomes; gene expression; infectious disease; microbiology.
© 2021, Anchimiuk et al.
Conflict of interest statement
AA, VL, FB, AM, FB, SG No competing interests declared
Figures




References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

