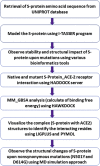
Investigation of nonsynonymous mutations in the spike protein of SARS-CoV-2 and its interaction with the ACE2 receptor by molecular docking and MM/GBSA approach
- PMID: 34346317
- PMCID: PMC8282961
- DOI: 10.1016/j.compbiomed.2021.104654
Investigation of nonsynonymous mutations in the spike protein of SARS-CoV-2 and its interaction with the ACE2 receptor by molecular docking and MM/GBSA approach
Abstract
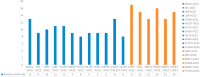
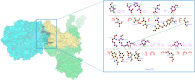
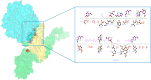
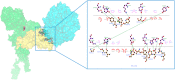
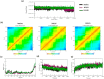
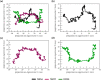
COVID-19 is an infectious and pathogenic viral disease caused by SARS-CoV-2 that leads to septic shock, coagulation dysfunction, and acute respiratory distress syndrome. The spreading rate of SARS-CoV-2 is higher than MERS-CoV and SARS-CoV. The receptor-binding domain (RBD) of the Spike-protein (S-protein) interacts with the human cells through the host angiotensin-converting enzyme 2 (ACE2) receptor. However, the molecular mechanism of pathological mutations of S-protein is still unclear. In this perspective, we investigated the impact of mutations in the S-protein and their interaction with the ACE2 receptor for SAR-CoV-2 viral infection. We examined the stability of pathological nonsynonymous mutations in the S-protein, and the binding behavior of the ACE2 receptor with the S-protein upon nonsynonymous mutations using the molecular docking and MM_GBSA approaches. Using the extensive bioinformatics pipeline, we screened the destabilizing (L8V, L8W, L18F, Y145H, M153T, F157S, G476S, L611F, A879S, C1247F, and C1254F) and stabilizing (H49Y, S50L, N501Y, D614G, A845V, and P1143L) nonsynonymous mutations in the S-protein. The docking and binding free energy (ddG) scores revealed that the stabilizing nonsynonymous mutations show increased interaction between the S-protein and the ACE2 receptor compared to native and destabilizing S-proteins and that they may have been responsible for the virulent high level. Further, the molecular dynamics simulation (MDS) approach reveals the structural transition of mutants (N501Y and D614G) S-protein. These insights might help researchers to understand the pathological mechanisms of the S-protein and provide clues regarding mutations in viral infection and disease propagation. Further, it helps researchers to develop an efficient treatment approach against this SARS-CoV-2 pandemic.
Keywords: ACE2 receptor; Binding affinity; Nonsynonymous mutations; SARS-CoV-2; Spike protein; Stability.
Copyright © 2021. Published by Elsevier Ltd.
Conflict of interest statement
We have no conflicts of interest to disclose.
Figures




References
-
- World Health Organization . World Health Organization; 2020. Coronavirus Disease 2019 (COVID-19): Situation Report; p. 52.https://apps.who.int/iris/handle/10665/331476
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

