Differential gene regulation in DAPT-treated Hydra reveals candidate direct Notch signalling targets
- PMID: 34346482
- PMCID: PMC8353520
- DOI: 10.1242/jcs.258768
Differential gene regulation in DAPT-treated Hydra reveals candidate direct Notch signalling targets
Abstract
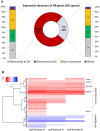
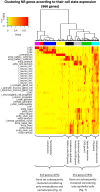
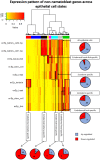
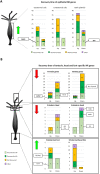
In Hydra, Notch inhibition causes defects in head patterning and prevents differentiation of proliferating nematocyte progenitor cells into mature nematocytes. To understand the molecular mechanisms by which the Notch pathway regulates these processes, we performed RNA-seq and identified genes that are differentially regulated in response to 48 h of treating the animals with the Notch inhibitor DAPT. To identify candidate direct regulators of Notch signalling, we profiled gene expression changes that occur during subsequent restoration of Notch activity and performed promoter analyses to identify RBPJ transcription factor-binding sites in the regulatory regions of Notch-responsive genes. Interrogating the available single-cell sequencing data set revealed the gene expression patterns of Notch-regulated Hydra genes. Through these analyses, a comprehensive picture of the molecular pathways regulated by Notch signalling in head patterning and in interstitial cell differentiation in Hydra emerged. As prime candidates for direct Notch target genes, in addition to Hydra (Hy)Hes, we suggest Sp5 and HyAlx. They rapidly recovered their expression levels after DAPT removal and possess Notch-responsive RBPJ transcription factor-binding sites in their regulatory regions.
Keywords: Axis formation; Hydra; Nematocyte differentiation; Notch pathway; Wnt pathway.
© 2021. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures




References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

