Mechanism and function of DNA replication-independent DNA-protein crosslink repair via the SUMO-RNF4 pathway
- PMID: 34346517
- PMCID: PMC8441304
- DOI: 10.15252/embj.2020107413
Mechanism and function of DNA replication-independent DNA-protein crosslink repair via the SUMO-RNF4 pathway
Abstract
DNA-protein crosslinks (DPCs) obstruct essential DNA transactions, posing a serious threat to genome stability and functionality. DPCs are proteolytically processed in a ubiquitin- and DNA replication-dependent manner by SPRTN and the proteasome but can also be resolved via targeted SUMOylation. However, the mechanistic basis of SUMO-mediated DPC resolution and its interplay with replication-coupled DPC repair remain unclear. Here, we show that the SUMO-targeted ubiquitin ligase RNF4 defines a major pathway for ubiquitylation and proteasomal clearance of SUMOylated DPCs in the absence of DNA replication. Importantly, SUMO modifications of DPCs neither stimulate nor inhibit their rapid DNA replication-coupled proteolysis. Instead, DPC SUMOylation provides a critical salvage mechanism to remove DPCs formed after DNA replication, as DPCs on duplex DNA do not activate interphase DNA damage checkpoints. Consequently, in the absence of the SUMO-RNF4 pathway cells are able to enter mitosis with a high load of unresolved DPCs, leading to defective chromosome segregation and cell death. Collectively, these findings provide mechanistic insights into SUMO-driven pathways underlying replication-independent DPC resolution and highlight their critical importance in maintaining chromosome stability and cellular fitness.
Keywords: DNA repair; DNA-protein crosslinks; SUMO; genome stability; ubiquitin.
© 2021 The Authors. Published under the terms of the CC BY 4.0 license.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
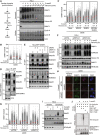
HeLa cells released from double thymidine synchronization in early S phase were treated with 5‐azadC for 30 min, washed and collected at the indicated times. Soluble and chromatin‐enriched fractions were immunoblotted with indicated antibodies.
U2OS cells treated as in (A) were pre‐extracted and immunostained with DNMT1 antibody. DNMT1 foci formation was analysed by quantitative image‐based cytometry (QIBC) (red bars, mean; > 1,500 cells analysed per condition). Data are representative of three independent experiments. See also Fig EV1A.
As in (B), except that cells were pre‐treated with the proteasome inhibitor MG132 and/or SUMOi for 30 and 15 min, respectively, before exposure to 5‐azadC (red bars, mean; > 1,900 cells analysed per condition). Data are representative of three independent experiments.
HeLa cells stably expressing GFP‐DNMT1 were treated or not with 5‐azadC for 30 min, collected and subjected to GFP immunoprecipitation under denaturing conditions, and immunoblotted with indicated antibodies.
HeLa/GFP‐DNMT1 cells transfected with indicated siRNAs were processed as in (D).
Immunoblot analysis of soluble and chromatin‐enriched fractions of HeLa cells transfected with indicated siRNAs, left untreated or exposed to 5‐azadC for 30 min and collected at the indicated times.
U2OS cells transfected with indicated siRNAs were treated with 5‐azadC for 30 min and processed for QIBC analysis of DNMT1 foci counts as in (B) (red bars, mean; > 5,600 cells analysed per condition). Data are representative of three independent experiments.
Representative images of U2OS cells transfected with indicated siRNAs followed by mCherry‐RNF4 expression plasmid. Cells were treated with 5‐azadC in the presence or absence of SUMOi, fixed 2 h later, pre‐extracted and co‐immunostained with DNMT1 and SUMO2/3 antibodies. Scale bar, 10 µm.
Immunoblot analysis of soluble and chromatin‐enriched fractions of HeLa cells treated with 5‐azadC and/or SUMOi as indicated.
GFP‐tagged DNMT1 from extracts of HeLa/GFP‐DNMT1 cells treated or not with 5‐azadC was immobilized on GFP‐Trap agarose, subjected to stringent washing to remove proteins non‐covalently bound to GFP‐DNMT1 and incubated with recombinant HA‐ubiquitin, E1 and E2 (UbcH5a) enzymes and RNF4 proteins (STUbL reaction) at 37°C for 1 h. Samples were then subjected to immunoblotting to assay for RNF4‐dependent STUbL activity towards GFP‐DNMT1.
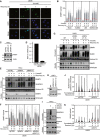
Representative images from the experiment shown in Fig 1B. Scale bar, 10 µm.
U2OS cells released from double thymidine synchronization in early S phase were pre‐treated or not with SUMOi for 15 min, pulse‐labelled with 5‐azadC for 30 min and incubated in the presence or absence of SUMOi and ubiquitin E1 inhibitor (UBi). Cells collected at the indicated times were pre‐extracted and immunostained with DNMT1 antibody. DNMT1 foci formation was analysed by quantitative image‐based cytometry (QIBC; red bars, mean; > 4,000 cells analysed per condition). Data are representative of three independent experiments.
Immunoblot analysis of HeLa cells transfected with non‐targeting control (CTRL) or RNF4 siRNA. *, cross‐reactive band.
U2OS cells were transfected with non‐targeting control (CTRL) or RNF111 siRNA. RNF111 mRNA levels were analysed by qPCR. Primers to GAPDH were used as a normalization control (mean ± SEM; n = 2 independent experiments).
HeLa cells transfected with indicated siRNAs were released from double thymidine synchronization in early S phase and treated with 5‐azadC for 30 min, washed and collected at the indicated times. Soluble and chromatin‐enriched fractions were immunoblotted with indicated antibodies.
U2OS cells transfected with indicated siRNAs were released from double thymidine block, pulse‐treated 2 h later with 5‐azadC for 30 min and processed for QIBC analysis of DNMT1 foci counts as in (B) (red bars, mean; > 8,500 cells analysed per condition). Data are representative of three independent experiments.
HAP1 cells (WT or independent derivative ΔRNF4 clones (#1–8 and #1–15) expressing a truncated form of RNF4 lacking the C‐terminal RING domain) were synchronized in S phase by overnight treatment with thymidine. Following removal of thymidine, cells were treated with 5‐azadC for 30 min and processed as in (E).
Immunoblot analysis of indicated HAP1 cell lines showing truncation and reduced expression of endogenous RNF4.
Immunoblot analysis of soluble and chromatin‐enriched fractions of HeLa cells treated with formaldehyde and/or SUMOi as indicated.
U2OS cells transfected with indicated siRNAs were treated with formaldehyde for 1 h. Cells were pre‐extracted and fixed at the indicated times after formaldehyde removal, immunostained with SUMO2/3 antibody and processed for QIBC analysis of SUMO2/3 foci counts (red bars, median; dashed lines, quartiles; > 6,000 cells analysed per condition). Data show SUMO2/3 foci counts per cell normalized to the mean foci count of untreated cells and are representative of three independent experiments.
As in (J), but showing total number of SUMO2/3 foci per cell.

M.HpaII was crosslinked into a plasmid to generate p4xDPC and then incubated in nucleoplasmic egg extracts (NPE) in the presence or absence of 50 µM of SUMOi. DPC pull‐down under stringent conditions was performed at the indicated time points, and the recovered samples were immunoblotted for crosslinked M.HpaII (Larsen et al, 2019). p4xDPC, which contains four M.HpaII DPCs, was used to increase sensitivity towards modified M.HpaII species. Identical results were obtained with plasmids containing one or four DPCs.
p4xDPC or p4xDPCSUMO (generated by crosslinking SUMOΔGG‐M.HpaII) were incubated in NPE and DPC pull‐down performed as in (A). Note that priming M.HpaII with SUMO stimulates rapid poly‐SUMOylation of the DPC in NPE. Thus, this substrate was used in many subsequent experiments to stimulate DPC poly‐SUMOylation.
p4xDPCSUMO was incubated in NPE and plasmids analysed as in (A). Following DPC pull‐down, the samples were split and either left untreated or treated with the SUMO protease Ulp1.
Sperm chromatin treated with 2,000 J/m2 of UV‐C was incubated in non‐replicating egg extracts in the presence or absence of the indicated PARP inhibitors (PARPi; Veliparib (50 µM), Olaparib (50 µM), Talazoparib (10 µM)). At the indicated time points, chromatin was recovered via chromatin spin‐down and samples were immunoblotted with indicated antibodies.
Plot illustrating protein recruitment to UV‐treated sperm chromatin in the presence or absence of Talazoparib and SUMOi, as determined by CHROMASS analysis (Dataset EV1). Red dots indicate the proteins that are significantly enriched on sperm chromatin in the presence of PARPi in a SUMO‐dependent manner (n = 4 biochemical replicates; FDR < 5% corresponds to a permutation‐based FDR‐adjusted q‐value of < 0.05). Note that different isoforms of the same protein (e.g. ATF7IP) can sometimes be detected.
NPE was either mock‐ or PIAS4‐depleted, and recombinant xPIAS4 was added where indicated to a final concentration of 10 ng/µl. Protein samples were immunoblotted with the indicated antibodies.
Samples from (F) were added to p4xDPCSUMO for the indicated times and recovered via DPC pull‐down as in (A).
pDPC or a plasmid containing M.HpaII crosslinked to ssDNA (pDPCssDNA) (Larsen et al, 2019) was incubated with SUMO E1 and E2 enzymes and SUMO, in the presence or absence of recombinant xPIAS4. Samples were recovered by DPC pull‐down and blotted against M.HpaII as in (A).
Representative images of U2OS cells transfected with Myc‐PIAS4 expression plasmid that were left untreated or exposed to 5‐azadC in the presence or absence of SUMOi, fixed 2 h later, pre‐extracted and co‐immunostained with DNMT1 and Myc antibodies. Scale bar, 5 µm.
HeLa or HeLa/GFP‐DNMT1 cells left untreated or exposed to 5‐azadC for 30 min were lysed and subjected to GFP IP under stringent conditions. After extensive washing, individual IPs were incubated with an equal amount (800 µg) of whole cell lysate of HeLa cells transfected with Myc‐PIAS4 expression construct (Fig EV2J), washed and immunoblotted with antibodies to Myc, SUMO2/3 and GFP. *, cross‐reactive band.
HeLa/GFP‐DNMT1 cells transfected with previously validated siRNAs targeting established SUMO E3 ligases (Fig EV2L and Methods section) were treated with 5‐azadC for 30 min, collected and subjected to GFP immunoprecipitation under denaturing conditions, and immunoblotted with antibodies to SUMO1, SUMO2/3, ubiquitin and GFP.
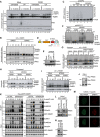
Sperm chromatin was left untreated or treated with 2,000 J/m2 of UV‐C and added to non‐replicating egg extracts in the presence or absence of 10 µM of Talazoparib (PARPi). At the indicated time points, chromatin was recovered via chromatin spin‐down and samples were immunoblotted with the indicated antibodies. Note that UV‐C triggers PARylation at 2 min (lane 7), which is abolished in the presence of PARPi.
Sperm chromatin was treated with 2,000 J/m2 of UV‐C and added to non‐replicating egg extracts in the presence of PARPi. SUMO E1 inhibitor was added where indicated. Chromatin was recovered via chromatin spin‐down and samples were immunoblotted with the indicated antibodies.
NPE was either mock‐ or PIAS4‐depleted, added to p4xDPC and analysed as in Fig 2A.
Schematic representation of PIAS4 functional domains and mutations introduced into the SIM motifs (Kaur et al, 2017).
NPE was either mock‐ or PIAS4‐depleted and supplemented with WT PIAS4 or a mutant containing inactivating substitutions in the SIM1 and SIM2 domains (SIM1/2*) (D). Protein samples were immunoblotted with the indicated antibodies.
Samples from (E) were added to p4xDPCSUMO and analysed as in Fig 2A.
Sperm chromatin was left untreated or exposed to 2,000 J/m2 of UV‐C and added to non‐replicating egg extracts that were either mock‐ or PIAS4‐depleted and supplemented with recombinant PIAS4 WT or SIM1/2*. PARPi was added where indicated. Chromatin was recovered via chromatin spin‐down, and samples were immunoblotted with the indicated antibodies.
Samples from Fig 2F were added to untreated sperm chromatin. At the indicated time points, chromatin was recovered via chromatin spin‐down and samples were immunoblotted with the indicated antibodies.
Samples from Fig 2F were added to UV‐C‐treated sperm chromatin in the presence of PARPi. At the indicated time points, chromatin was recovered via chromatin spin‐down and samples were immunoblotted with the indicated antibodies.
Immunoblot analysis of whole cell lysate of HeLa cells transfected or not with Myc‐PIAS4 expression construct. Lysate from Myc‐PIAS4‐expressing cells was distributed equally between the three individual IP conditions in Fig 2J (800 µg per sample).
HeLa/GFP‐DNMT1 cells transfected with non‐targeting control (CTRL) or PIAS4 siRNAs were treated with 5‐azadC for the indicated times, collected and subjected to GFP immunoprecipitation under denaturing conditions, and immunoblotted with indicated antibodies.
Immunoblot analysis of HeLa/GFP‐DNMT1 cells transfected with non‐targeting control (CTRL) or PIAS4 siRNAs.
U2OS cells were transfected with control (CTRL) or PIAS4 siRNA targeting the 3′UTR and subsequently transfected with plasmids encoding WT or catalytically inactive (CI) Myc‐PIAS4 expression plasmid. Cells were then exposed to 5‐azadC, fixed 1 h later and co‐immunostained with Myc and DNMT1 antibodies. Representative images are shown. Note that Myc‐PIAS4 CI is recruited to DNMT1 DPCs even in the absence of endogenous PIAS4 activity. Scale bar, 5 µm.
Immunoblot analysis of U2OS cells transfected with control (CTRL) or PIAS4 siRNA targeting the 3′UTR.

Scheme illustrating replication in egg extracts of p2xDPCLeads, a plasmid containing two M.HpaII crosslinked on opposite strands. Under these conditions, the vast majority of replication forks encounter the DPC on their leading strand template (Larsen et al, 2019).
p2xDPCLeads or p2xDPCSUMOLeads were replicated in egg extracts in the presence [α‐32P]dATP. Where indicated, 50 µM of SUMOi was added to the extracts. Reaction samples were analysed by native agarose gel electrophoresis. RI, replication intermediates; OC, open circular; SC, supercoiled. Red arrowheads indicate OC molecules that have not yet undergone repair.
Samples in (B) were recovered by DPC pull‐down and immunoblotted for crosslinked M.HpaII.
Scheme illustrating sequential SUMOylation and ubiquitylation of DPC‐containing plasmids in non‐replicating egg extracts. First, p4xDPCSUMO is incubated in NPE for 60‐90 min to achieve poly‐SUMOylation of the DPCs. NPE is then supplemented with recombinant RNF4 to a final concentration of 7 ng/µl (E) or an equal volume of CSF‐arrested whole egg extract (F, G) to trigger DPC ubiquitylation.
p4xDPCSUMO was incubated in NPE for 60 min and supplemented with buffer or recombinant RNF4 (WT or a catalytically inactive C130A mutant). At the indicated time points following RNF4 addition, the DPC plasmid was recovered by DPC pull‐down and immunoblotted against M.HpaII.
pDPCSUMO was subjected to sequential extract addition as depicted in (D). Recombinant UBC9DN was added to NPE where indicated to block de novo SUMOylation. At the indicated time points following addition of CSF‐arrested egg extract, the plasmid was recovered via DPC pull‐down and immunoblotted against M.HpaII.
pDPCSUMO was subjected to sequential extract addition as depicted in (D). Ninety min after addition of CSF‐arrested extract, samples were recovered via DPC pull‐down and treated with the SUMO protease Ulp1 or the ubiquitin protease Usp2 as indicated. Samples were then immunoblotted against M.HpaII.
CSF‐arrested extracts were either mock‐ or RNF4‐depleted and recombinant His‐RNF4 was supplemented to RNF4‐depleted extracts to a final concentration of 7 ng/µl where indicated. Protein samples were immunoblotted with the indicated antibodies.
p4xDPCSUMO was polySUMOylated in NPE, recovered via DPC pull‐down and incubated in fresh CSF‐arrested extract that was either mock‐ or RNF4‐depleted. At indicated time points following CSF extract addition, the plasmid was recovered and immunoblotted against M.HpaII.
As in (I), but using RNF4‐depleted CSF‐arrested extracts reconstituted with recombinant RNF4 WT or C130A from (H).
CSF‐arrested extracts were either mock‐ or PSMA1‐depleted. Protein samples were immunoblotted with the indicated antibodies.
p4xDPCSUMO was subjected to sequential addition of NPE and CSF‐arrested extract as in (I). CSF‐arrested extracts from (K) were supplemented with recombinant RNF4, and the DPC plasmid was recovered by DPC pull‐down at the indicated time points and immunoblotted against M.HpaII.

- A
p2xDPCLeads was replicated in egg extracts in the presence of radiolabelled nucleotides in the presence or absence of the SUMO E1 or ubiquitin E1 inhibitors. Samples were analysed by native agarose gel electrophoresis as in Fig 3B. Red arrowheads indicate open circular molecules (OC) that have not yet undergone repair.
- B
Samples from (A) were recovered by DPC pull‐down and immunoblotted against M.HpaII. Note that in the presence of the ubiquitin E1 inhibitor DPC ubiquitylation and degradation by both SPRTN and the proteasome are severely inhibited (Duxin et al, ; Larsen et al, 2019). Residual ubiquitylation likely reflects ubiquitin E1 enzymes in the extract activated prior to addition of the inhibitor.
- C
Scheme illustrating DPC repair in egg extracts when M.HpaII or SUMO‐M.HpaII is crosslinked to ssDNA (pDPCssDNA or pDPCSUMOssDNA). In this setting, DPC ubiquitylation is mainly driven by the E3 ubiquitin ligase RFWD3 and does not require DNA replication (Gallina et al, 2021).
- D
pDPCssDNA or pDPCSUMO‐ssDNA were incubated in non‐replicating egg extracts. DPCs were recovered by pull‐down and immunoblotted against M.HpaII. Note the similar kinetics of degradation for SUMOylated and non‐SUMOylated DPCs.
- E
Indicated volumes of whole egg CSF‐arrested extract and NPE were immunoblotted with RNF4 antibody next to a dilution series of recombinant His‐RNF4. A band migrating around 28 kDa and immunodepleted with the RNF4 antibody (Fig 3H) is indicated as RNF4. * denotes non‐specific bands that are not immunodepleted by the RNF4 antibody.
- F
Samples from Fig 3E were immunoblotted with the indicated antibodies.
- G
p4xDPCSUMO was incubated in NPE or CSF‐arrested extracts. At the indicated times, the DPCs were recovered and immunoblotted against M.HpaII.
- H
Indicated volumes of whole egg CSF‐arrested extract and NPE were immunoblotted with PIAS4 and PSMA3 antibodies. Where indicated, CSF‐arrested extract was supplemented with 10 ng/µl of recombinant PIAS4.
- I
p4xDPCSUMO was incubated with CSF‐arrested extracts from (H). DPCs were recovered at the indicated time points and immunoblotted against M.HpaII.
- J
p4xDPCSUMO was incubated in NPE for 60 min and supplemented with buffer or either 7 ng/µl or 35 ng/µl (5×) of recombinant RNF4 (WT) in the presence of MG262 where indicated. Protein samples were immunoblotted with the indicated antibodies.
- K
Samples from (J) were recovered via DPC pull‐down at the indicated time following RNF4 addition and immunoblotted against M.HpaII.
- L, M
Independent replicates of Fig 3L.
- N
Sperm chromatin was incubated for 1 h in CSF‐arrested egg extract in the presence or absence of 10 µM of the pan‐CDK inhibitor R547. Note that chromosome condensation is inhibited in the presence of R547, consistent with CDK inhibition. Scale bar, 30 µm.
- O
Sequential addition of Xenopus egg extracts to DPC‐containing plasmids. After the first addition of NPE, the SUMOylated DPCs were recovered and incubated in CSF‐arrested egg extract in the presence or absence of R547. The DPCs were again recovered and immunoblotted against M.HpaII.

HeLa cells were transfected with indicated siRNAs and synchronized in early S phase by double thymidine block. Following release from the block, cells were pulse‐labelled with 5‐azadC in late S phase for 30 min. Cells were then collected at the indicated times after 5‐azadC withdrawal and analysed by flow cytometry. Data are representative of three independent experiments. Proportion of cells with G2/M DNA content is indicated.
RPE1 cells were synchronized in early S phase by double thymidine block. Following release from the block, cells were pulse‐labelled or not with 5‐azadC for 30 min in late S phase, incubated with nocodazole, collected at the indicated times and immunoblotted with indicated antibodies.
As in (B), except that SUMOi was added to the culture medium 15 min prior to 5‐azadC treatment.
As in (B), except that cells were transfected with RNF4 siRNA prior to double thymidine block synchronization.
Immunoblot analysis of RPE1 cells exposed to indicated genotoxic agents and collected 1 h later.
Immunoblot analysis of HeLa cells exposed to indicated genotoxic agents and collected 1 h later.
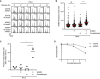
HeLa cells transfected with indicated siRNAs were treated or not with 5‐azadC for 30 min and/or MPS1i in late S phase. Cells were then collected at the indicated times after 5‐azadC withdrawal and analysed by flow cytometry. Data are representative of three independent experiments. Proportion of cells with G2/M DNA content is indicated.
HeLa cells were synchronized in early S phase by double thymidine block, released and pulse‐treated 7 h later with formaldehyde for 1 h in the presence or absence of SUMOi. Following formaldehyde removal, cells were subjected to live‐cell imaging analysis, and the duration of mitosis (nuclear envelope breakdown (NEBD) to anaphase onset) was quantified (red bars, median; at least 156 cells, pooled from three independent experiments, were analysed per condition; ****P < 0.0001, ns: not significant, Mann–Whitney test).
Quantification of mitotic defects in cells in (B) (black bars, mean; n = 3 independent experiments; > 150 cells quantified per condition; *P < 0.05, paired t‐test).
Clonogenic survival of HeLa cells transfected with indicated siRNAs and subjected to indicated doses of formaldehyde for 30 min before replating (mean ± SEM; n = 2 independent experiments).

Outline of experimental set up to monitor DNMT1 DPC levels in S phase and mitotic cells.
HeLa cells released from double thymidine block in early S phase were mock‐treated or pulse‐labelled for 30 min with 5‐azadC in the presence of SUMOi in late S phase. Following 5‐azadC removal, cells were incubated with SUMOi and nocodazole, and mitotic cells were collected by shake‐off, as outlined in (A). Soluble and chromatin‐enriched fractions were immunoblotted with indicated antibodies.
U2OS cells transfected with indicated siRNAs and then treated as in (B) were collected in late S phase or mitosis (M), subjected to stringent pre‐extraction and immunostained with DNMT1 antibody. DNMT1 foci formation was quantified by QIBC analysis (red bars, mean; > 7,400 cells analysed per condition). Data are representative of three independent experiments.
HeLa cells transfected with indicated siRNAs and synchronized in early S phase by double thymidine block were pulse‐labelled with 5‐azadC in late S phase as outlined in (A). Cells were then collected at the indicated times after 5‐azadC withdrawal and analysed by flow cytometry. Data are representative of three independent experiments. Proportion of cells with G2/M DNA content is indicated.
HeLa cells transfected with indicated siRNAs were treated with 5‐azadC and/or SUMOi in late S phase as outlined in (A). Cells were then collected at the indicated times after 5‐azadC withdrawal and analysed by flow cytometry. Data are representative of three independent experiments. Proportion of cells with G2/M DNA content is indicated.
HeLa cells transfected with indicated siRNAs were pulse‐labelled or not with 5‐azadC for 30 min in late S phase according to the experimental setup in (A). Following 5‐azadC withdrawal cells were incubated with nocodazole, collected at the indicated times and immunoblotted with indicated antibodies.
HeLa cells transfected or not with indicated siRNAs were synchronized in early S phase by double thymidine block, released and pulse‐labelled 5,5 h later with 5‐azadC for 30 min in the presence or absence of SUMOi. Following 5‐azadC removal, cells were subjected to live‐cell imaging analysis, and the duration from late S phase to mitotic entry (nuclear envelope breakdown (NEBD)) was quantified (red bars, median; at least 83 cells, pooled from three independent experiments, were analysed per condition; **P < 0.01, ns: not significant, Mann–Whitney test).
Immunoblot analysis of siRNA‐treated HeLa cells that were exposed to indicated genotoxic agents and collected 1 h later.
- A
HeLa cells transfected or not with indicated siRNAs were synchronized in early S phase by double thymidine block, released and pulse‐labelled 5,5 h later with 5‐azadC for 30 min in the presence or absence of SUMOi. Following 5‐azadC removal, cells were subjected to live‐cell imaging analysis, and the duration of mitosis (nuclear envelope breakdown (NEBD) to anaphase onset) was quantified (red bars, median; at least 72 cells, pooled from three independent experiments, were analysed per condition; ****P < 0.0001, ns: not significant, Mann–Whitney test).
- B
Representative time‐lapse sequences of mitotic progression in cells in (A). NEBD corresponds to t = 0. White arrowheads indicate cells undergoing division. Scale bar, 10 µm.
- C, D
Quantification of mitotic defects in cells in (A) (mean; n = 3 independent experiments; > 97 cells quantified per condition; *P < 0.05, **P < 0.01, paired t‐test).
- E
HeLa cells transfected with RNF4 siRNA were synchronized in early S phase by double thymidine block. Six hours after release from the block, cells were pulse‐labelled with 5‐azadC for 30 min. Mitotic cells were isolated by shake‐off 8 h later, washed and incubated or not with MPS1 inhibitor (MPS1i) and collected at the indicated times. Cells were then processed for immunoblotting with indicated antibodies.
- F
Clonogenic survival of 5‐azadC‐treated HeLa cells transfected with indicated siRNAs (mean ± SEM; n = 2 independent experiments).
- G
Clonogenic survival of 5‐azadC‐treated WT HAP1 cells and derivative cell lines expressing a truncated form of RNF4 lacking the RING domain (ΔRNF4; Fig EV1H) (mean ± SEM; n = 2 independent experiments).

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

