Variant PNLDC1, Defective piRNA Processing, and Azoospermia
- PMID: 34347949
- PMCID: PMC7615015
- DOI: 10.1056/NEJMoa2028973
Variant PNLDC1, Defective piRNA Processing, and Azoospermia
Abstract
Background: P-element-induced wimpy testis (PIWI)-interacting RNAs (piRNAs) are short (21 to 35 nucleotides in length) and noncoding and are found almost exclusively in germ cells, where they regulate aberrant expression of transposable elements and postmeiotic gene expression. Critical to the processing of piRNAs is the protein poly(A)-specific RNase-like domain containing 1 (PNLDC1), which trims their 3' ends and, when disrupted in mice, causes azoospermia and male infertility.
Methods: We performed exome sequencing on DNA samples from 924 men who had received a diagnosis of nonobstructive azoospermia. Testicular-biopsy samples were analyzed by means of histologic and immunohistochemical tests, in situ hybridization, reverse-transcriptase-quantitative-polymerase-chain-reaction assay, and small-RNA sequencing.
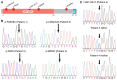
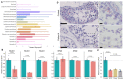
Results: Four unrelated men of Middle Eastern descent who had nonobstructive azoospermia were found to carry mutations in PNLDC1: the first patient had a biallelic stop-gain mutation, p.R452Ter (rs200629089; minor allele frequency, 0.00004); the second, a novel biallelic missense variant, p.P84S; the third, two compound heterozygous mutations consisting of p.M259T (rs141903829; minor allele frequency, 0.0007) and p.L35PfsTer3 (rs754159168; minor allele frequency, 0.00004); and the fourth, a novel biallelic canonical splice acceptor site variant, c.607-2A→T. Testicular histologic findings consistently showed error-prone meiosis and spermatogenic arrest with round spermatids of type Sa as the most advanced population of germ cells. Gene and protein expression of PNLDC1, as well as the piRNA-processing proteins PIWIL1, PIWIL4, MYBL1, and TDRKH, were greatly diminished in cells of the testes. Furthermore, the length distribution of piRNAs and the number of pachytene piRNAs was significantly altered in men carrying PNLDC1 mutations.
Conclusions: Our results suggest a direct mechanistic effect of faulty piRNA processing on meiosis and spermatogenesis in men, ultimately leading to male infertility. (Funded by Innovation Fund Denmark and others.).
Copyright © 2021 Massachusetts Medical Society.
Figures


Comment in
-
Defective piRNA Processing and Azoospermia.N Engl J Med. 2022 Apr 28;386(17):1674-1675. doi: 10.1056/NEJMc2116008. N Engl J Med. 2022. PMID: 35476664 No abstract available.
-
Defective piRNA Processing and Azoospermia.N Engl J Med. 2022 Apr 28;386(17):1675. doi: 10.1056/NEJMc2116008. N Engl J Med. 2022. PMID: 35476665 No abstract available.
-
Defective piRNA Processing and Azoospermia.N Engl J Med. 2022 Apr 28;386(17):1675-1676. doi: 10.1056/NEJMc2116008. N Engl J Med. 2022. PMID: 35476666 No abstract available.
References
-
- Kasak L, Laan M. Monogenic causes of non-obstructive azoospermia: challenges, established knowledge, limitations and perspectives. Hum Genet. 2021;140:135–54. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
