SARS-CoV-2 Variants in Patients with Immunosuppression
- PMID: 34347959
- PMCID: PMC8494465
- DOI: 10.1056/NEJMsb2104756
SARS-CoV-2 Variants in Patients with Immunosuppression
Figures
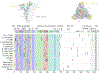

References
-
- Truong TT, Ryutov A, Pandey U, et al. Persistent SARS-CoV-2 infection and increasing viral variants in children and young adults with impaired humoral immunity. March 2, 2021. (https://www.medrxiv.org/content/10.1101/2021.02.27.21252099v1). preprint. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
