Apicidin biosynthesis is linked to accessory chromosomes in Fusarium poae isolates
- PMID: 34348672
- PMCID: PMC8340494
- DOI: 10.1186/s12864-021-07617-y
Apicidin biosynthesis is linked to accessory chromosomes in Fusarium poae isolates
Abstract
Background: Fusarium head blight is a disease of global concern that reduces crop yields and renders grains unfit for consumption due to mycotoxin contamination. Fusarium poae is frequently associated with cereal crops showing symptoms of Fusarium head blight. While previous studies have shown F. poae isolates produce a range of known mycotoxins, including type A and B trichothecenes, fusarins and beauvericin, genomic analysis suggests that this species may have lineage-specific accessory chromosomes with secondary metabolite biosynthetic gene clusters awaiting description.
Methods: We examined the biosynthetic potential of 38 F. poae isolates from Eastern Canada using a combination of long-read and short-read genome sequencing and untargeted, high resolution mass spectrometry metabolome analysis of extracts from isolates cultured in multiple media conditions.
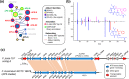
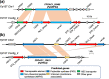
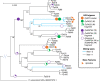
Results: A high-quality assembly of isolate DAOMC 252244 (Fp157) contained four core chromosomes as well as seven additional contigs with traits associated with accessory chromosomes. One of the predicted accessory contigs harbours a functional biosynthetic gene cluster containing homologs of all genes associated with the production of apicidins. Metabolomic and genomic analyses confirm apicidins are produced in 4 of the 38 isolates investigated and genomic PCR screening detected the apicidin synthetase gene APS1 in approximately 7% of Eastern Canadian isolates surveyed.
Conclusions: Apicidin biosynthesis is linked to isolate-specific putative accessory chromosomes in F. poae. The data produced here are an important resource for furthering our understanding of accessory chromosome evolution and the biosynthetic potential of F. poae.
Keywords: Accessory chromosomes; Apicidin; Biosynthetic gene clusters; Fungal plant pathogens; Fusarium poae; Genomics; Mass spectrometry; Metabolomics; Secondary metabolites.
© 2021. Her Majesty the Queen in Right of Canada as represented by the Minister of Agriculture and Agri-Food.
Conflict of interest statement
The authors declare they have no competing interests.
Figures


Similar articles
-
Naturally Occurring Fusarium Species and Mycotoxins in Oat Grains from Manitoba, Canada.Toxins (Basel). 2021 Sep 18;13(9):670. doi: 10.3390/toxins13090670. Toxins (Basel). 2021. PMID: 34564673 Free PMC article.
-
Development of a PCR assay to detect the potential production of nivalenol in Fusarium poae.FEMS Microbiol Lett. 2012 Jul;332(2):99-104. doi: 10.1111/j.1574-6968.2012.02581.x. Epub 2012 May 15. FEMS Microbiol Lett. 2012. PMID: 22536946
-
Diversity in metabolite production by Fusarium langsethiae, Fusarium poae, and Fusarium sporotrichioides.Int J Food Microbiol. 2004 Sep 15;95(3):257-66. doi: 10.1016/j.ijfoodmicro.2003.12.005. Int J Food Microbiol. 2004. PMID: 15337591
-
Accessory Chromosome-Acquired Secondary Metabolism in Plant Pathogenic Fungi: The Evolution of Biotrophs Into Host-Specific Pathogens.Front Microbiol. 2021 Apr 23;12:664276. doi: 10.3389/fmicb.2021.664276. eCollection 2021. Front Microbiol. 2021. PMID: 33968000 Free PMC article. Review.
-
Molecular biology of Fusarium mycotoxins.Int J Food Microbiol. 2007 Oct 20;119(1-2):47-50. doi: 10.1016/j.ijfoodmicro.2007.07.024. Epub 2007 Jul 31. Int J Food Microbiol. 2007. PMID: 17707105 Review.
Cited by
-
Debunking the Myth of Fusarium poae T-2/HT-2 Toxin Production.J Agric Food Chem. 2024 Feb 28;72(8):3949-3957. doi: 10.1021/acs.jafc.3c08437. Epub 2024 Feb 20. J Agric Food Chem. 2024. PMID: 38375818 Free PMC article.
-
Poplar leaf bud resin metabolomics: seasonal profiling of leaf bud chemistry in Populus trichocarpa provides insight into resin biosynthesis.Plant Cell Physiol. 2025 Mar 31;66(3):291-303. doi: 10.1093/pcp/pcae149. Plant Cell Physiol. 2025. PMID: 39699046 Free PMC article.
-
Pathogenicity and Metabolomic Characterization of Fusarium graminearum and Fusarium poae Challenge in Barley under Controlled Conditions.J Fungi (Basel). 2024 Sep 26;10(10):670. doi: 10.3390/jof10100670. J Fungi (Basel). 2024. PMID: 39452622 Free PMC article.
-
Identification and Confirmation of Virulence Factor Production from Fusarium avenaceum, a Causal Agent of Root Rot in Pulses.J Fungi (Basel). 2024 Nov 26;10(12):821. doi: 10.3390/jof10120821. J Fungi (Basel). 2024. PMID: 39728317 Free PMC article.
-
Digital PCR assays for quantifying trichothecene-producing Fusarium species, including Fusarium langsethiae, F. poae, and F. sporotrichioides, in oats.Anal Bioanal Chem. 2025 May;417(13):2957-2969. doi: 10.1007/s00216-025-05840-0. Epub 2025 Mar 21. Anal Bioanal Chem. 2025. PMID: 40116867 Free PMC article.
References
-
- Chakraborty S, Newton AC. Climate change, plant diseases and food security: an overview. Plant Pathol. 2011;60(1):2–14. doi: 10.1111/j.1365-3059.2010.02411.x. - DOI
-
- Vogelgsang S, Beyer M, Pasquali M, Jenny E, Musa T, Bucheli TD, Wettstein FE, Forrer HR. An eight-year survey of wheat shows distinctive effects of cropping factors on different Fusarium species and associated mycotoxins. Eur J Agron. 2019;105:62–77. doi: 10.1016/j.eja.2019.01.002. - DOI
-
- Xue AG, Chen Y, Seifert K, Guo W, Blackwell BA, Harris LJ, et al. Prevalence of Fusarium species causing head blight of spring wheat, barley and oat in Ontario during 2001–2017. Can J Plant Pathol 2019;0(00):1–11.
-
- Xu XM, Parry DW, Nicholson P, Thomsett MA, Simpson D, Edwards SG, et al. Predominance and association of pathogenic fungi causing Fusarium ear blightin wheat in four European countries. Eur J Plant Pathol. 2005;112(2):143–154. doi: 10.1007/s10658-005-2446-7. - DOI
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources

