Pathogen Challenge and Dietary Shift Alter Microbiota Composition and Activity in a Mucin-Associated in vitro Model of the Piglet Colon (MPigut-IVM) Simulating Weaning Transition
- PMID: 34349744
- PMCID: PMC8328230
- DOI: 10.3389/fmicb.2021.703421
Pathogen Challenge and Dietary Shift Alter Microbiota Composition and Activity in a Mucin-Associated in vitro Model of the Piglet Colon (MPigut-IVM) Simulating Weaning Transition
Abstract
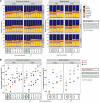
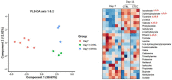
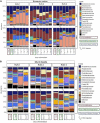
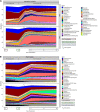
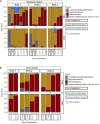
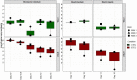
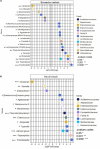
Enterotoxigenic Escherichia coli (ETEC) is the principal pathogen responsible for post-weaning diarrhea in newly weaned piglets. Expansion of ETEC at weaning is thought to be the consequence of various stress factors such as transient anorexia, dietary change or increase in intestinal inflammation and permeability, but the exact mechanisms remain to be elucidated. As the use of animal experiments raise more and more ethical concerns, we used a recently developed in vitro model of piglet colonic microbiome and mucobiome, the MPigut-IVM, to evaluate the effects of a simulated weaning transition and pathogen challenge at weaning. Our data suggested that the tested factors impacted the composition and functionality of the MPigut-IVM microbiota. The simulation of weaning transition led to an increase in relative abundance of the Prevotellaceae family which was further promoted by the presence of the ETEC strain. In contrast, several beneficial families such as Bacteroidiaceae or Ruminococcaceae and gut health related short chain fatty acids like butyrate or acetate were reduced upon simulated weaning. Moreover, the incubation of MPigut-IVM filtrated effluents with porcine intestinal cell cultures showed that ETEC challenge in the in vitro model led to an increased expression of pro-inflammatory genes by the porcine cells. This study provides insights about the etiology of a dysbiotic microbiota in post-weaning piglets.
Keywords: ETEC; gene expression; in vitro model of colonic microbiota; intestinal cells; piglet; weaning.
Copyright © 2021 Gresse, Chaucheyras-Durand, Garrido, Denis, Jiménez-Marín, Beaumont, Van de Wiele, Forano and Blanquet-Diot.
Conflict of interest statement
FC-D and RG are employees of Lallemand SAS. The authors declare that this study received funding from Lallemand SAS. The funder had the following involvement in the study: study design, data analysis, interpretation of the data and writing of the article. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Barba-Vidal E., Castillejos L., López-Colom P., Rivero Urgell M., Moreno Muñoz J. A., Martín-Orúe S. M. (2017). Evaluation of the probiotic strain Bifidobacterium longum subsp. Infantis CECT 7210 capacities to improve health status and fight digestive pathogens in a piglet model. Front. Microbiol. 8:533. 10.3389/fmicb.2017.00533 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

