Analysis methods and quality criteria for investigating muscle physiology using x-ray diffraction
- PMID: 34351359
- PMCID: PMC8348228
- DOI: 10.1085/jgp.202012778
Analysis methods and quality criteria for investigating muscle physiology using x-ray diffraction
Abstract
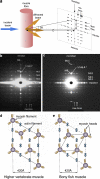
X-ray diffraction studies of muscle have been tremendously powerful in providing fundamental insights into the structures of, for example, the myosin and actin filaments in a variety of muscles and the physiology of the cross-bridge mechanism during the contractile cycle. However, interpretation of x-ray diffraction patterns is far from trivial, and if modeling of the observed diffraction intensities is required it needs to be performed carefully with full knowledge of the possible pitfalls. Here, we discuss (1) how x-ray diffraction can be used as a tool to monitor various specific muscle properties and (2) how to get the most out of the rest of the observed muscle x-ray diffraction patterns by modeling where the reliability of the modeling conclusions can be objectively tested. In other x-ray diffraction methods, such as protein crystallography, the reliability of every step of the process is estimated and quoted in published papers. In this way, the quality of the structure determination can be properly assessed. To be honest with ourselves in the muscle field, we need to do as near to the same as we can, within the limitations of the techniques that we are using. We discuss how this can be done. We also use test cases to reveal the dos and don'ts of using x-ray diffraction to study muscle physiology.
© 2021 Squire and Knupp.
Figures







Comment in
-
Myosin motors that cannot bind actin leave their folded OFF state on activation of skeletal muscle.J Gen Physiol. 2021 Nov 1;153(11):e202112896. doi: 10.1085/jgp.202112896. Epub 2021 Oct 20. J Gen Physiol. 2021. PMID: 34668926 Free PMC article.
References
-
- AL-Khayat, H.A., Hudson L., Reedy M.K., Irving T.C., and Squire J.M.. 2003. Myosin head configuration in relaxed insect flight muscle: x-ray modeled resting cross-bridges in a pre-powerstroke state are poised for actin binding. Biophys. J. 85:1063–1079. 10.1016/S0006-3495(03)74545-7 - DOI - PMC - PubMed

