A ubiquitin-like protein encoded by the "noncoding" RNA TINCR promotes keratinocyte proliferation and wound healing
- PMID: 34351912
- PMCID: PMC8341662
- DOI: 10.1371/journal.pgen.1009686
A ubiquitin-like protein encoded by the "noncoding" RNA TINCR promotes keratinocyte proliferation and wound healing
Abstract
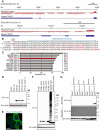
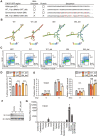
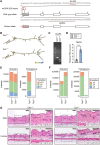
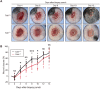
Although long noncoding RNAs (lncRNAs) are transcripts that do not encode proteins by definition, some lncRNAs actually contain small open reading frames that are translated. TINCR (terminal differentiation-induced ncRNA) has been recognized as a lncRNA that contributes to keratinocyte differentiation. However, we here show that TINCR encodes a ubiquitin-like protein that is well conserved among species and whose expression was confirmed by the generation of mice harboring a FLAG epitope tag sequence in the endogenous open reading frame as well as by targeted proteomics. Forced expression of this protein promoted cell cycle progression in normal human epidermal keratinocytes, and mice lacking this protein manifested a delay in skin wound healing associated with attenuated cell cycle progression in keratinocytes. We termed this protein TINCR-encoded ubiquitin-like protein (TUBL), and our results reveal a role for TINCR in the regulation of keratinocyte proliferation and skin regeneration that is dependent on TUBL.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

