A simple model explains the cell cycle-dependent assembly of centromeric nucleosomes in holocentric species
- PMID: 34352103
- PMCID: PMC8450114
- DOI: 10.1093/nar/gkab648
A simple model explains the cell cycle-dependent assembly of centromeric nucleosomes in holocentric species
Abstract
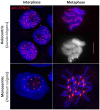
Centromeres are essential for chromosome movement. In independent taxa, species with holocentric chromosomes exist. In contrast to monocentric species, where no obvious dispersion of centromeres occurs during interphase, the organization of holocentromeres differs between condensed and decondensed chromosomes. During interphase, centromeres are dispersed into a large number of CENH3-positive nucleosome clusters in a number of holocentric species. With the onset of chromosome condensation, the centromeric nucleosomes join and form line-like holocentromeres. Using polymer simulations, we propose a mechanism relying on the interaction between centromeric nucleosomes and structural maintenance of chromosomes (SMC) proteins. Different sets of molecular dynamic simulations were evaluated by testing four parameters: (i) the concentration of Loop Extruders (LEs) corresponding to SMCs, (ii) the distribution and number of centromeric nucleosomes, (iii) the effect of centromeric nucleosomes on interacting LEs and (iv) the assembly of kinetochores bound to centromeric nucleosomes. We observed the formation of a line-like holocentromere, due to the aggregation of the centromeric nucleosomes when the chromosome was compacted into loops. A groove-like holocentromere structure formed after a kinetochore complex was simulated along the centromeric line. Similar mechanisms may also organize a monocentric chromosome constriction, and its regulation may cause different centromere types during evolution.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
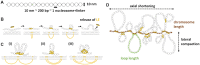




References
-
- Melters D.P., Paliulis L.V., Korf I.F., Chan S.W.L.. Holocentric chromosomes: convergent evolution, meiotic adaptations, and genomic analysis. Chromosome Res. 2012; 20:579–593. - PubMed
-
- Escudero M., Mrquez-Corro J.I., Hipp A.L.. The phylogenetic origins and evolutionary history of holocentric chromosomes. Syst. Bot. 2016; 41:580–585.
-
- Král J., Forman M., Kořínková T., Lerma A.C.R., Haddad C.R., Musilová J., Řezáč M., Herrera I.M.Á., Thakur S., Dippenaar-Schoeman A.S.et al.. Insights into the karyotype and genome evolution of haplogyne spiders indicate a polyploid origin of lineage with holokinetic chromosomes. Sci. Rep.-uk. 2019; 9:3001. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

