De novo assembly, annotation, and comparative analysis of 26 diverse maize genomes
- PMID: 34353948
- PMCID: PMC8733867
- DOI: 10.1126/science.abg5289
De novo assembly, annotation, and comparative analysis of 26 diverse maize genomes
Abstract
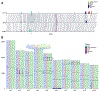
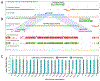
We report de novo genome assemblies, transcriptomes, annotations, and methylomes for the 26 inbreds that serve as the founders for the maize nested association mapping population. The number of pan-genes in these diverse genomes exceeds 103,000, with approximately a third found across all genotypes. The results demonstrate that the ancient tetraploid character of maize continues to degrade by fractionation to the present day. Excellent contiguity over repeat arrays and complete annotation of centromeres revealed additional variation in major cytological landmarks. We show that combining structural variation with single-nucleotide polymorphisms can improve the power of quantitative mapping studies. We also document variation at the level of DNA methylation and demonstrate that unmethylated regions are enriched for cis-regulatory elements that contribute to phenotypic variation.
Copyright © 2021, American Association for the Advancement of Science.
Figures


References
-
- Hirsch CN, Hirsch CD, Brohammer AB, Bowman MJ, Soifer I, Barad O, Shem-Tov D, Baruch K, Lu F, Hernandez AG, Fields CJ, Wright CL, Koehler K, Springer NM, Buckler E, Buell CR, de Leon N, Kaeppler SM, Childs KL, Mikel MA, Draft Assembly of Elite Inbred Line PH207 Provides Insights into Genomic and Transcriptome Diversity in Maize. Plant Cell. 28, 2700–2714 (2016). - PMC - PubMed
-
- Ricci WA, Lu Z, Ji L, Marand AP, Ethridge CL, Murphy NG, Noshay JM, Galli M, Mejía-Guerra MK, Colomé-Tatché M, Johannes F, Rowley MJ, Corces VG, Zhai J, Scanlon MJ, Buckler ES, Gallavotti A, Springer NM, Schmitz RJ, Zhang X, Widespread long-range cis-regulatory elements in the maize genome. Nature Plants 5, 1237–1249 (2019). - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

