Diverse heterochromatin-associated proteins repress distinct classes of genes and repetitive elements
- PMID: 34354237
- PMCID: PMC9248069
- DOI: 10.1038/s41556-021-00725-7
Diverse heterochromatin-associated proteins repress distinct classes of genes and repetitive elements
Erratum in
-
Author Correction: Diverse heterochromatin-associated proteins repress distinct classes of genes and repetitive elements.Nat Cell Biol. 2021 Nov;23(11):1212. doi: 10.1038/s41556-021-00759-x. Nat Cell Biol. 2021. PMID: 34588637 No abstract available.
Abstract
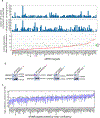
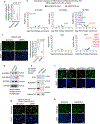
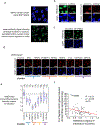
Heterochromatin, typically marked by histone H3 trimethylation at lysine 9 (H3K9me3) or lysine 27 (H3K27me3), represses different protein-coding genes in different cells, as well as repetitive elements. The basis for locus specificity is unclear. Previously, we identified 172 proteins that are embedded in sonication-resistant heterochromatin (srHC) harbouring H3K9me3. Here, we investigate in humans how 97 of the H3K9me3-srHC proteins repress heterochromatic genes. We reveal four groups of srHC proteins that each repress many common genes and repeat elements. Two groups repress H3K9me3-embedded genes with different extents of flanking srHC, one group is specific for srHC genes with H3K9me3 and H3K27me3, and one group is specific for genes with srHC as the primary feature. We find that the enhancer of rudimentary homologue (ERH) is conserved from Schizosaccharomyces pombe in repressing meiotic genes and, in humans, now represses other lineage-specific genes and repeat elements. The study greatly expands our understanding of H3K9me3-based gene repression in vertebrates.
© 2021. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing Interests Statement
The authors have no competing interests to declare.
Figures













Comment in
-
Deconfining heterochromatin for expression.Nat Cell Biol. 2021 Aug;23(8):814-816. doi: 10.1038/s41556-021-00726-6. Nat Cell Biol. 2021. PMID: 34354235 No abstract available.
References
-
- Bulut-Karslioglu A. et al. Suv39h-dependent H3K9me3 marks intact retrotransposons and silences LINE elements in mouse embryonic stem cells. Mol. Cell 55, 277–290 (2014). - PubMed
References for Methods
-
- van Pelt JF, Decorte R, Yap PSH & Fevery J. Identification of HepG2 variant cell lines by short tandem repeat (STR) analysis. Mol Cell Biochem 243, 49–54 (2003). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

