ICTV Virus Taxonomy Profile: Marnaviridae 2021
- PMID: 34356002
- PMCID: PMC8513639
- DOI: 10.1099/jgv.0.001633
ICTV Virus Taxonomy Profile: Marnaviridae 2021
Abstract
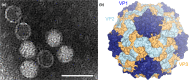
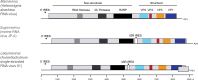
The family Marnaviridae comprises small non-enveloped viruses with positive-sense RNA genomes of 8.6-9.6 kb. Isolates infect marine single-celled eukaryotes (protists) that come from diverse lineages. Some members are known from metagenomic studies of ocean virioplankton, with additional unclassified viruses described from metagenomic datasets derived from marine and freshwater environments. This is a summary of the International Committee on Taxonomy of Viruses (ICTV) Report on the family Marnaviridae, which is available at ictv.global/report/marnaviridae.
Keywords: ICTV Report; Marnaviridae; taxonomy.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures
References
-
- Tai V, Lawrence JE, Lang AS, Chan AM, Culley AI, et al. Characterization of HaRNAV, a single‐stranded RNA virus causing lysis of Heterosigma akashiwo (Raphidophyceae) . J Phycol. 2003;39:343–352.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

