Mating-Type Locus Organization and Mating-Type Chromosome Differentiation in the Bipolar Edible Button Mushroom Agaricus bisporus
- PMID: 34356095
- PMCID: PMC8305134
- DOI: 10.3390/genes12071079
Mating-Type Locus Organization and Mating-Type Chromosome Differentiation in the Bipolar Edible Button Mushroom Agaricus bisporus
Abstract
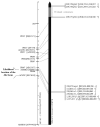
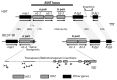
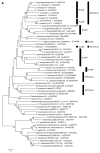
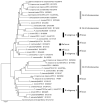
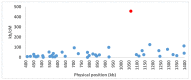
In heterothallic basidiomycete fungi, sexual compatibility is restricted by mating types, typically controlled by two loci: PR, encoding pheromone precursors and pheromone receptors, and HD, encoding two types of homeodomain transcription factors. We analysed the single mating-type locus of the commercial button mushroom variety, Agaricus bisporus var. bisporus, and of the related variety burnettii. We identified the location of the mating-type locus using genetic map and genome information, corresponding to the HD locus, the PR locus having lost its mating-type role. We found the mip1 and β-fg genes flanking the HD genes as in several Agaricomycetes, two copies of the β-fg gene, an additional HD2 copy in the reference genome of A. bisporus var. bisporus and an additional HD1 copy in the reference genome of A. bisporus var. burnettii. We detected a 140 kb-long inversion between mating types in an A. bisporus var. burnettii heterokaryon, trapping the HD genes, the mip1 gene and fragments of additional genes. The two varieties had islands of transposable elements at the mating-type locus, spanning 35 kb in the A. bisporus var. burnettii reference genome. Linkage analyses showed a region with low recombination in the mating-type locus region in the A. bisporus var. burnettii variety. We found high differentiation between β-fg alleles in both varieties, indicating an ancient event of recombination suppression, followed more recently by a suppression of recombination at the mip1 gene through the inversion in A. bisporus var. burnettii and a suppression of recombination across whole chromosomes in A. bisporus var. bisporus, constituting stepwise recombination suppression as in many other mating-type chromosomes and sex chromosomes.
Keywords: Agaricus; automixis; basidiomycete; bipolar; genetic map; heterothallic; homeodomain genes; mating-type chromosome; mating-type genes; pheromone genes; pheromone receptor genes; pseudo-homothallic; recombination suppression; sex chromosomes; tetrapolar.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses or interpretation of data; in the writing of the manuscript or in the decision to publish the results.
Figures

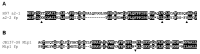




Similar articles
-
An expanded genetic linkage map of an intervarietal Agaricus bisporus var. bisporusxA. bisporus var. burnettii hybrid based on AFLP, SSR and CAPS markers sheds light on the recombination behaviour of the species.Fungal Genet Biol. 2010 Mar;47(3):226-36. doi: 10.1016/j.fgb.2009.12.003. Epub 2009 Dec 21. Fungal Genet Biol. 2010. PMID: 20026415
-
Telomere-to-telomere assembled and centromere annotated genomes of the two main subspecies of the button mushroom Agaricus bisporus reveal especially polymorphic chromosome ends.Sci Rep. 2020 Sep 4;10(1):14653. doi: 10.1038/s41598-020-71043-5. Sci Rep. 2020. PMID: 32887908 Free PMC article.
-
Conservation of genetic linkage with map expansion in distantly related crosses of Agaricus bisporus.FEMS Microbiol Lett. 1997 Jan 15;146(2):235-40. doi: 10.1111/j.1574-6968.1997.tb10199.x. FEMS Microbiol Lett. 1997. PMID: 9011044
-
Recombination suppression and evolutionary strata around mating-type loci in fungi: documenting patterns and understanding evolutionary and mechanistic causes.New Phytol. 2021 Mar;229(5):2470-2491. doi: 10.1111/nph.17039. Epub 2020 Dec 1. New Phytol. 2021. PMID: 33113229 Free PMC article. Review.
-
Mating-type genes for basidiomycete strain improvement in mushroom farming.Appl Microbiol Biotechnol. 2001 Sep;56(5-6):602-12. doi: 10.1007/s002530100763. Appl Microbiol Biotechnol. 2001. PMID: 11601606 Review.
Cited by
-
Whole-Genome Sequencing and Comparative Genomics Analysis of the Wild Edible Mushroom (Gomphus purpuraceus) Provide Insights into Its Potential Food Application and Artificial Domestication.Genes (Basel). 2022 Sep 10;13(9):1628. doi: 10.3390/genes13091628. Genes (Basel). 2022. PMID: 36140797 Free PMC article. Review.
-
Stepwise recombination suppression around the mating-type locus in an ascomycete fungus with self-fertile spores.PLoS Genet. 2023 Feb 10;19(2):e1010347. doi: 10.1371/journal.pgen.1010347. eCollection 2023 Feb. PLoS Genet. 2023. PMID: 36763677 Free PMC article.
-
Chromosome-Wide Characterization of Intragenic Crossover in Shiitake Mushroom, Lentinula edodes.J Fungi (Basel). 2021 Dec 15;7(12):1076. doi: 10.3390/jof7121076. J Fungi (Basel). 2021. PMID: 34947058 Free PMC article.
-
Lessons on fruiting body morphogenesis from genomes and transcriptomes of Agaricomycetes.Stud Mycol. 2023 Jul;104:1-85. doi: 10.3114/sim.2022.104.01. Epub 2023 Jan 31. Stud Mycol. 2023. PMID: 37351542 Free PMC article.
-
Transcription factors: switches for regulating growth and development in macrofungi.Appl Microbiol Biotechnol. 2023 Oct;107(20):6179-6191. doi: 10.1007/s00253-023-12726-7. Epub 2023 Aug 25. Appl Microbiol Biotechnol. 2023. PMID: 37624406 Review.
References
-
- Charlesworth D., Charlesworth B. Inbreeding depression and its evolutionary consequences. Annu. Rev. Ecol. Syst. 1987;18:237–268. doi: 10.1146/annurev.es.18.110187.001321. - DOI
-
- Igic B., Lande R., Kohn J. Loss of self-incompatibility and its evolutionary consequences. Int. J. Plant Sci. 2008;169:93–104. doi: 10.1086/523362. - DOI
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

