How Important Are Structural Variants for Speciation?
- PMID: 34356100
- PMCID: PMC8305853
- DOI: 10.3390/genes12071084
How Important Are Structural Variants for Speciation?
Abstract
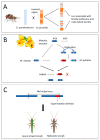
Understanding the genetic basis of reproductive isolation is a central issue in the study of speciation. Structural variants (SVs); that is, structural changes in DNA, including inversions, translocations, insertions, deletions, and duplications, are common in a broad range of organisms and have been hypothesized to play a central role in speciation. Recent advances in molecular and statistical methods have identified structural variants, especially inversions, underlying ecologically important traits; thus, suggesting these mutations contribute to adaptation. However, the contribution of structural variants to reproductive isolation between species-and the underlying mechanism by which structural variants most often contribute to speciation-remain unclear. Here, we review (i) different mechanisms by which structural variants can generate or maintain reproductive isolation; (ii) patterns expected with these different mechanisms; and (iii) relevant empirical examples of each. We also summarize the available sequencing and bioinformatic methods to detect structural variants. Lastly, we suggest empirical approaches and new research directions to help obtain a more complete assessment of the role of structural variants in speciation.
Keywords: hybridization; reproductive isolation; suppressed recombination.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures


References
-
- Wu C.-I. The genic view of the process of speciation. J. Evol. Biol. 2001;14:851–865. doi: 10.1046/j.1420-9101.2001.00335.x. - DOI
-
- Coyne J.A., Orr H.A. Speciation. Volume 37. Sinauer; Sunderland, MA, USA: 2004.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

