Plasmonic Interferometers as TREM2 Sensors for Alzheimer's Disease
- PMID: 34356688
- PMCID: PMC8301914
- DOI: 10.3390/bios11070217
Plasmonic Interferometers as TREM2 Sensors for Alzheimer's Disease
Abstract
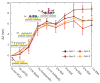
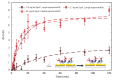
We report an effective surface immobilization protocol for capture of Triggering Receptor Expressed on Myeloid Cells 2 (TREM2), a receptor whose elevated concentration in cerebrospinal fluid has recently been associated with Alzheimer's disease (AD). We employ the proposed surface functionalization scheme to design, fabricate, and assess a biochemical sensing platform based on plasmonic interferometry that is able to detect physiological concentrations of TREM2 in solution. These findings open up opportunities for label-free biosensing of TREM2 in its soluble form in various bodily fluids as an early indicator of the onset of clinical dementia in AD. We also show that plasmonic interferometry can be a powerful tool to monitor and optimize surface immobilization schemes, which could be applied to develop other relevant antibody tests.
Keywords: Alzheimer’s disease; TREM2 sensors; optical biosensor; plasmonic interferometry; surface functionalization.
Conflict of interest statement
The authors declare that there are no conflicts of interest related to this article.
Figures




Similar articles
-
TREM2 ectodomain and its soluble form in Alzheimer's disease.J Neuroinflammation. 2020 Jul 7;17(1):204. doi: 10.1186/s12974-020-01878-2. J Neuroinflammation. 2020. PMID: 32635934 Free PMC article. Review.
-
Discovery and engineering of an anti-TREM2 antibody to promote amyloid plaque clearance by microglia in 5XFAD mice.MAbs. 2022 Jan-Dec;14(1):2107971. doi: 10.1080/19420862.2022.2107971. MAbs. 2022. PMID: 35921534 Free PMC article.
-
TREM2 regulates innate immunity in Alzheimer's disease.J Neuroinflammation. 2018 Apr 14;15(1):107. doi: 10.1186/s12974-018-1148-y. J Neuroinflammation. 2018. PMID: 29655369 Free PMC article. Review.
-
Disease Progression-Dependent Effects of TREM2 Deficiency in a Mouse Model of Alzheimer's Disease.J Neurosci. 2017 Jan 18;37(3):637-647. doi: 10.1523/JNEUROSCI.2110-16.2016. J Neurosci. 2017. PMID: 28100745 Free PMC article.
-
TREM2 Expression and Amyloid-Beta Phagocytosis in Alzheimer's Disease.Int J Mol Sci. 2023 May 11;24(10):8626. doi: 10.3390/ijms24108626. Int J Mol Sci. 2023. PMID: 37239970 Free PMC article.
Cited by
-
Unlocking early detection of Alzheimer's disease: The emerging role of nanomaterial-based optical sensors.J Food Drug Anal. 2024 Sep 13;32(3):296-324. doi: 10.38212/2224-6614.3520. J Food Drug Anal. 2024. PMID: 39636776 Free PMC article. Review.
References
-
- Vos T., Allen C., Arora M., Barber R.M., Bhutta Z.A., Brown A., Carter A., Casey D.C., Charlson F.J., Chen A.Z., et al. Global, regional, and national incidence, prevalence, and years lived with disability for 310 diseases and injuries, 1990–2015: A systematic analysis for the Global Burden of Disease Study 2015. Lancet. 2016;388:1545–1602. doi: 10.1016/S0140-6736(16)31678-6. - DOI - PMC - PubMed
-
- World Health Organization . Towards a Dementia Plan: A WHO Guide. World Health Organization; Geneva, Switzerland: 2018.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

