Different Within-Host Viral Evolution Dynamics in Severely Immunosuppressed Cases with Persistent SARS-CoV-2
- PMID: 34356872
- PMCID: PMC8301427
- DOI: 10.3390/biomedicines9070808
Different Within-Host Viral Evolution Dynamics in Severely Immunosuppressed Cases with Persistent SARS-CoV-2
Abstract
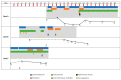
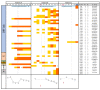
A successful Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) variant, B.1.1.7, has recently been reported in the UK, causing global alarm. Most likely, the new variant emerged in a persistently infected patient, justifying a special focus on these cases. Our aim in this study was to explore certain clinical profiles involving severe immunosuppression that may help explain the prolonged persistence of viable viruses. We present three severely immunosuppressed cases (A, B, and C) with a history of lymphoma and prolonged SARS-CoV-2 shedding (2, 4, and 6 months), two of whom finally died. Whole-genome sequencing of 9 and 10 specimens from Cases A and B revealed extensive within-patient acquisition of diversity, 12 and 28 new single nucleotide polymorphisms, respectively, which suggests ongoing SARS-CoV-2 replication. This diversity was not observed for Case C after analysing 5 sequential nasopharyngeal specimens and one plasma specimen, and was only observed in one bronchoaspirate specimen, although viral viability was still considered based on constant low Ct values throughout the disease and recovery of the virus in cell cultures. The acquired viral diversity in Cases A and B followed different dynamics. For Case A, new single nucleotide polymorphisms were quickly fixed (13-15 days) after emerging as minority variants, while for Case B, higher diversity was observed at a slower emergence: fixation pace (1-2 months). Slower SARS-CoV-2 evolutionary pace was observed for Case A following the administration of hyperimmune plasma. This work adds knowledge on SARS-CoV-2 prolonged shedding in severely immunocompromised patients and demonstrates viral viability, noteworthy acquired intra-patient diversity, and different SARS-CoV-2 evolutionary dynamics in persistent cases.
Keywords: COVID-19; SARS-CoV-2; diversity; genomics; immunosuppressed; persistence; viral viability.
Conflict of interest statement
The authors declare no conflict of interests.
Figures
References
-
- Alm E., Broberg E.K., Connor T., Hodcroft E.B., Komissarov A.B., Maurer-Stroh S., Melidou A., Neher R.A., O’Toole A., Pereyaslov D. Geographical and temporal distribution of SARS-CoV-2 clades in the WHO European Region, January to June 2020. Eurosurveillance. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.32.2001410. - DOI - PMC - PubMed
-
- Hodcroft E.B., Zuber M., Nadeau S., Crawford K.H.D., Bloom J.D., Veesler D., Vaughan T.G., Comas I., Candelas F.G., Stadler T., et al. Emergence and spread of a SARS-CoV-2 variant through Europe in the summer of 2020. Medrxiv Prepr. Serv. Health Sci. 2020 doi: 10.1101/2020.10.25.20219063. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

