Do Spectra Live in the Matrix? A Brief Tutorial on Applications of Factor Analysis to Resolving Spectral Datasets of Mixtures
- PMID: 34357495
- PMCID: PMC8547214
- DOI: 10.1007/s10895-021-02753-w
Do Spectra Live in the Matrix? A Brief Tutorial on Applications of Factor Analysis to Resolving Spectral Datasets of Mixtures
Abstract
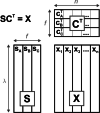
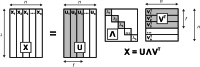
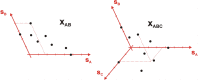
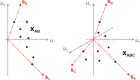
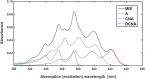
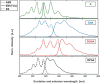
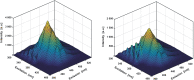
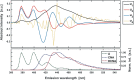
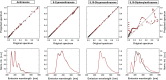
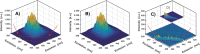
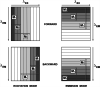
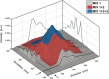
In spite of a rapid growth of data processing software, that has allowed for a huge advancement in many fields of chemistry, some research issues still remain problematic. A standard example of a troublesome challenge is the analysis of multi-component mixtures. The classical approach to such a problem consists of separating each component from a sample and performing individual measurements. The advent of computers, however, gave rise to a relatively new domain of data processing - chemometry - focused on decomposing signal recorded for the sample rather than the sample itself. Regrettably, still a very few chemometric methods are practically used in everyday laboratory routines. The Authors believe that a brief 'user-friendly' guide-like article on several 'flagship' algorithms of chemometrics may, at least partly, stimulate an increased interest in the use of these techniques among researchers specializing in many fields of chemistry. In the paper, five different techniques of factor analysis are used for the analysis of a three-component system of fluorophores. These algorithms, applied on the excitation-emission spectra, recorded for the 'unknown' mixture, allowed to unambiguously determine its composition without the need for physical separation of the components. An example of using chemometric methods for physical chemistry research is also provided. For each presented technique of the data analysis, a short description of its theoretical background followed by an example of its practical performance is given. In addition, the Reader is supplemented with a basic information on matrix algebra, detailed experimental 'recipes', reference specialist literature and ready-to-use MATLAB codes.
Keywords: Evolving factor analysis; Excitation-emission maps; Fluorescence quenching; Multivariate curve resolution; Rank annihilation factor analysis; Spectral data matrices of mixtures.
© 2021. The Author(s).
Conflict of interest statement
The authors have no conflicts of interest to declare that are relevant to the content of this article.
Figures



References
-
- Zinatloo-Ajabshir S, Heidari-Asil SA, Salavati-Niasari M. Simple and eco-friendly synthesis of recoverable zinc cobalt oxide-based ceramic nanostructure as high-performance photocatalyst for enhanced photocatalytic removal of organic contamination under solar light. Sep Purif Technol. 2021;267:118667. doi: 10.1016/j.seppur.2021.118667. - DOI
-
- Rubio-Clemente A, Chica E, Peñuela GA. Rapid determination of anthracene and benzo (a) pyrene by high-performance liquid chromatography with fluorescence detection. Anal Lett. 2017;50:1229–1247. doi: 10.1080/00032719.2016.1225304. - DOI
-
- Nie S, Dadoo R, Zare RN. Ultrasensitive fluorescence detection of polycyclic aromatic hydrocarbons in capillary electrophoresis. Anal Chem. 1993;65:3571–3575. doi: 10.1021/ac00072a007. - DOI
-
- Malinowski ER, Howery DG. Factor analysis in chemistry. New York: Wiley; 1980.
-
- Maeder M, Neuhold YM. Practical data analysis in chemistry. Amsterdam: Elsevier; 2007.
LinkOut - more resources
Full Text Sources

