Accessing Dietary Effects on the Rumen Microbiome: Different Sequencing Methods Tell Different Stories
- PMID: 34357930
- PMCID: PMC8310016
- DOI: 10.3390/vetsci8070138
Accessing Dietary Effects on the Rumen Microbiome: Different Sequencing Methods Tell Different Stories
Abstract
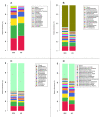
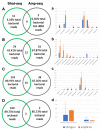
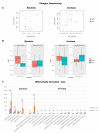
The current study employed both amplicon and shotgun sequencing to examine and compare the rumen microbiome in Angus bulls fed with either a backgrounding diet (BCK) or finishing diet (HG), to assess if both methods produce comparable results. Rumen digesta samples from 16 bulls were subjected for microbial profiling. Distinctive microbial profiles were revealed by the two methods, indicating that choice of sequencing approach may be a critical facet in studies of the rumen microbiome. Shotgun-sequencing identified the presence of 303 bacterial genera and 171 archaeal species, several of which exhibited differential abundance. Amplicon-sequencing identified 48 bacterial genera, 4 archaeal species, and 9 protozoal species. Among them, 20 bacterial genera and 5 protozoal species were differentially abundant between the two diets. Overall, amplicon-sequencing showed a more drastic diet-derived effect on the ruminal microbial profile compared to shotgun-sequencing. While both methods detected dietary differences at various taxonomic levels, few consistent patterns were evident. Opposite results were seen for the phyla Firmicutes and Bacteroidetes, and the genus Selenomonas. This study showcases the importance of sequencing platform choice and suggests a need for integrative methods that allow robust comparisons of microbial data drawn from various omic approaches, allowing for comprehensive comparisons across studies.
Keywords: amplicon-sequencing; bull cattle; diets; rumen; shotgun-sequencing.
Conflict of interest statement
The authors declare that this study was performed in the absence of any relationships or influences that could be construed as a conflict of interest.
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources

